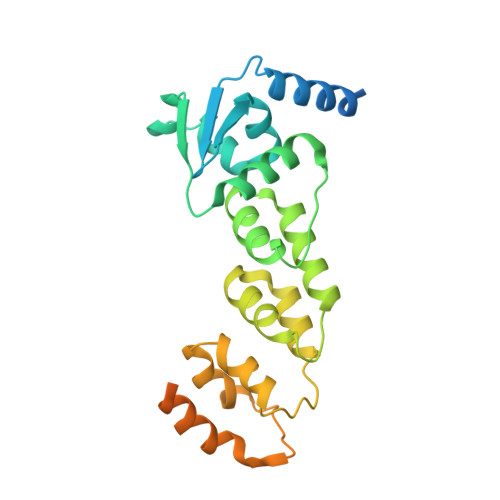
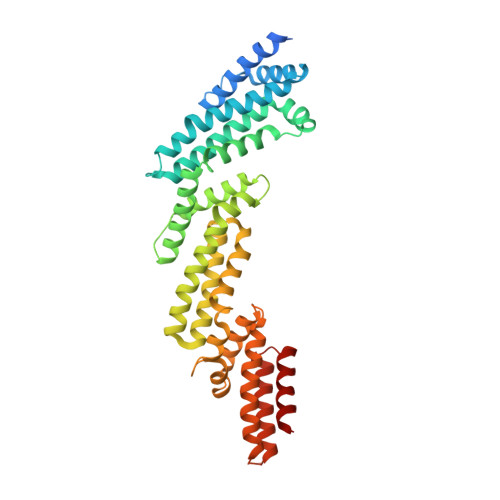
Crystal structure of KLHL3 in complex with Cullin3.
Ji, A.X., Prive, G.G.(2013) PLoS One 8: e60445-e60445
- PubMed: 23573258
- DOI: https://doi.org/10.1371/journal.pone.0060445
- Primary Citation of Related Structures:
4HXI - PubMed Abstract:
KLHL3 is a BTB-BACK-Kelch family protein that serves as a substrate adapter in Cullin3 (Cul3) E3 ubiquitin ligase complexes. KLHL3 is highly expressed in distal nephron tubules where it is involved in the regulation of electrolyte homeostasis and blood pressure. Mutations in KLHL3 have been identified in patients with inherited hypertension disorders, and several of the disease-associated mutations are located in the presumed Cul3 binding region. Here, we report the crystal structure of a complex between the KLHL3 BTB-BACK domain dimer and two copies of an N terminal fragment of Cul3. We use isothermal titration calorimetry to directly demonstrate that several of the disease mutations in the KLHL3 BTB-BACK domains disrupt the association with Cul3. Both the BTB and BACK domains contribute to the Cul3 interaction surface, and an extended model of the dimeric CRL3 complex places the two E2 binding sites in a suprafacial arrangement with respect to the presumed substrate-binding sites.
Organizational Affiliation:
Department of Biochemistry, University of Toronto, Toronto, Ontario, Canada.