Structural Basis for Recruitment and Activation of the AP-1 Clathrin Adaptor Complex by Arf1.
Ren, X., Farias, G.G., Canagarajah, B.J., Bonifacino, J.S., Hurley, J.H.(2013) Cell 152: 755-767
- PubMed: 23415225
- DOI: https://doi.org/10.1016/j.cell.2012.12.042
- Primary Citation of Related Structures:
4HMY - PubMed Abstract:
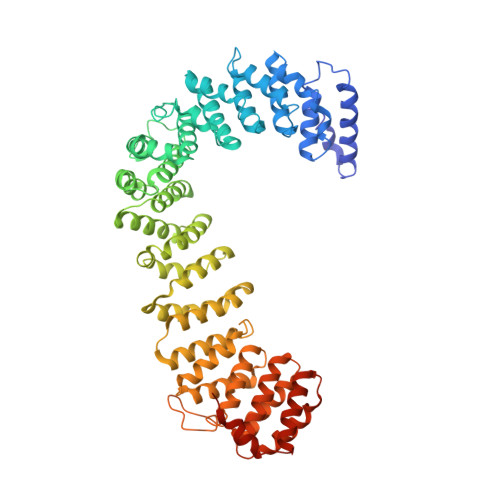
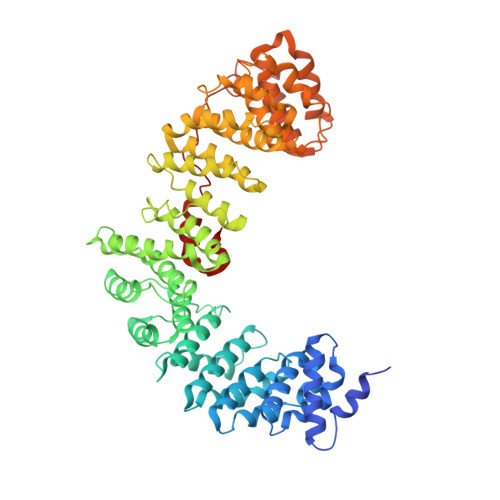
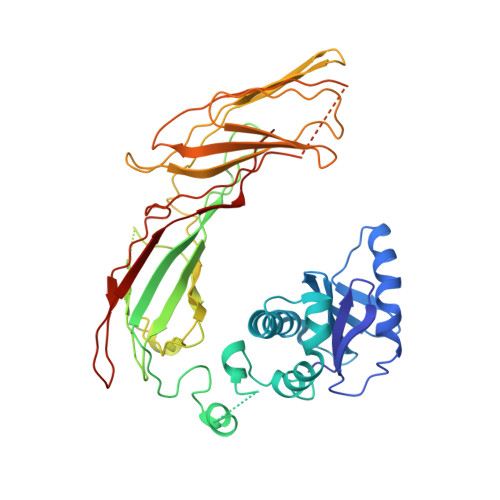
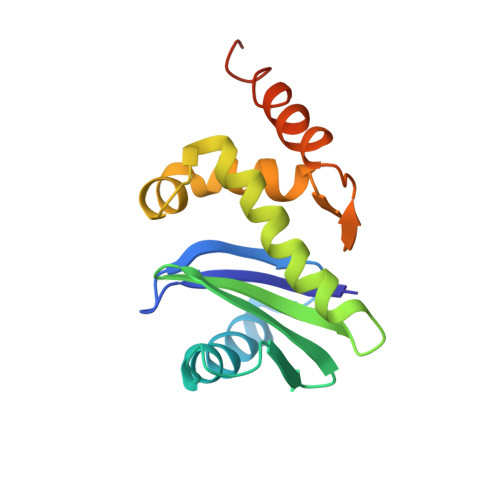
AP-1 is a clathrin adaptor complex that sorts cargo between the trans-Golgi network and endosomes. AP-1 recruitment to these compartments requires Arf1-GTP. The crystal structure of the tetrameric core of AP-1 in complex with Arf1-GTP, together with biochemical analyses, shows that Arf1 activates cargo binding by unlocking AP-1. Unlocking is driven by two molecules of Arf1 that bridge two copies of AP-1 at two interaction sites. The GTP-dependent switch I and II regions of Arf1 bind to the N terminus of the β1 subunit of one AP-1 complex, while the back side of Arf1 binds to the central part of the γ subunit trunk of a second AP-1 complex. A third Arf1 interaction site near the N terminus of the γ subunit is important for recruitment, but not activation. These observations lead to a model for the recruitment and activation of AP-1 by Arf1.
Organizational Affiliation:
Cell Biology and Metabolism Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA.