Long-range Electrostatic Complementarity Governs Substrate Recognition by Human Chymotrypsin C, a Key Regulator of Digestive Enzyme Activation.
Batra, J., Szabo, A., Caulfield, T.R., Soares, A.S., Sahin-Toth, M., Radisky, E.S.(2013) J Biol Chem 288: 9848-9859
- PubMed: 23430245
- DOI: https://doi.org/10.1074/jbc.M113.457382
- Primary Citation of Related Structures:
4H4F - PubMed Abstract:
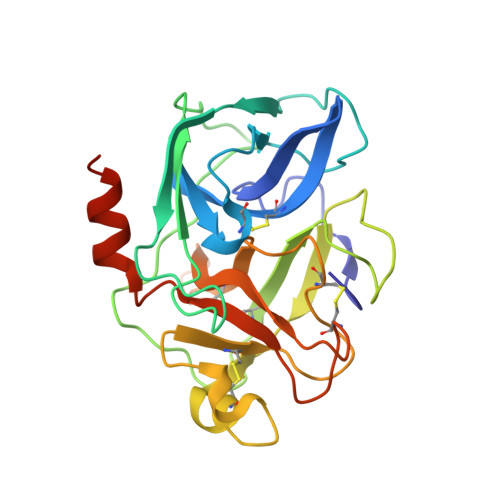
Human chymotrypsin C (CTRC) is a pancreatic serine protease that regulates activation and degradation of trypsinogens and procarboxypeptidases by targeting specific cleavage sites within their zymogen precursors. In cleaving these regulatory sites, which are characterized by multiple flanking acidic residues, CTRC shows substrate specificity that is distinct from that of other isoforms of chymotrypsin and elastase. Here, we report the first crystal structure of active CTRC, determined at 1.9-Å resolution, revealing the structural basis for binding specificity. The structure shows human CTRC bound to the small protein protease inhibitor eglin c, which binds in a substrate-like manner filling the S6-S5' subsites of the substrate binding cleft. Significant binding affinity derives from burial of preferred hydrophobic residues at the P1, P4, and P2' positions of CTRC, although acidic P2' residues can also be accommodated by formation of an interfacial salt bridge. Acidic residues may also be specifically accommodated in the P6 position. The most unique structural feature of CTRC is a ring of intense positive electrostatic surface potential surrounding the primarily hydrophobic substrate binding site. Our results indicate that long-range electrostatic attraction toward substrates of concentrated negative charge governs substrate discrimination, which explains CTRC selectivity in regulating active digestive enzyme levels.
Organizational Affiliation:
Department of Cancer Biology, Mayo Clinic Cancer Center, Jacksonville, Florida 32224.