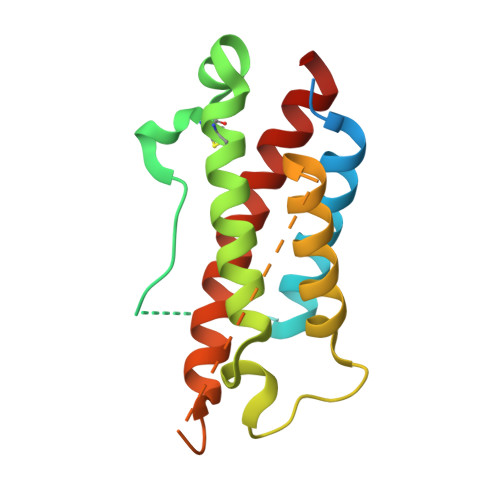
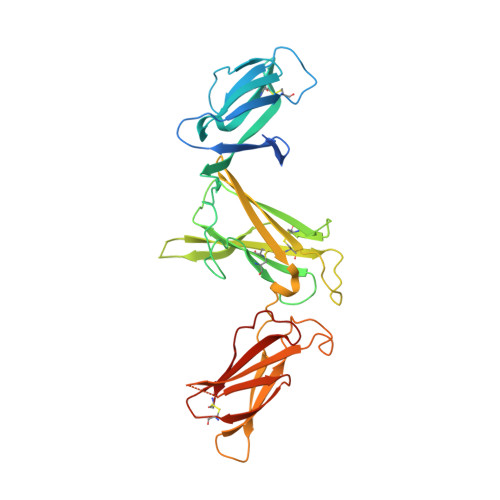
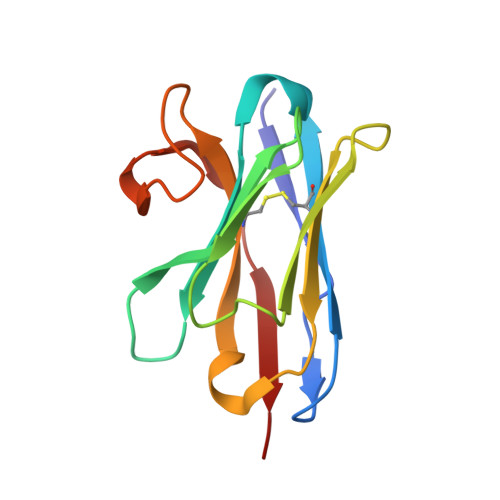
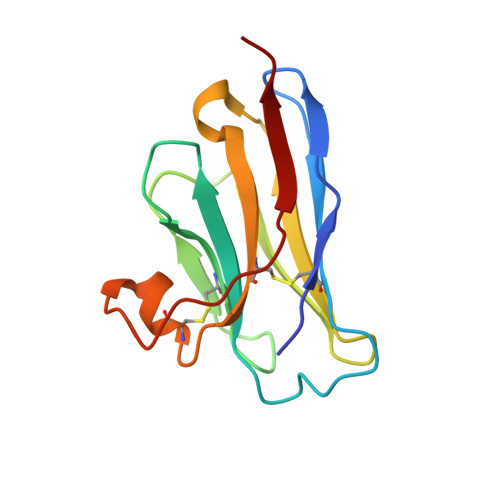
Neutralization of Human Interleukin 23 by Multivalent Nanobodies Explained by the Structure of Cytokine-Nanobody Complex.
Desmyter, A., Spinelli, S., Boutton, C., Saunders, M., Blachetot, C., de Haard, H., Denecker, G., Van Roy, M., Cambillau, C., Rommelaere, H.(2017) Front Immunol 8: 884-884
- PubMed: 28871249
- DOI: https://doi.org/10.3389/fimmu.2017.00884
- Primary Citation of Related Structures:
4GRW - PubMed Abstract:
The heterodimeric cytokine interleukin (IL) 23 comprises the IL12-shared p40 subunit and an IL23-specific subunit, p19. Together with IL12 and IL27, IL23 sits at the apex of the regulatory mechanisms shaping adaptive immune responses. IL23, together with IL17, plays an important role in the development of chronic inflammation and autoimmune inflammatory diseases. In this context, we generated monovalent antihuman IL23 variable heavy chain domain of llama heavy chain antibody (V HH ) domains (Nanobodies ® ) with low nanomolar affinity for human interleukin (hIL) 23. The crystal structure of a quaternary complex assembling hIL23 and several nanobodies against p19 and p40 subunits allowed identification of distinct epitopes and enabled rational design of a multivalent IL23-specific blocking nanobody. Taking advantage of the ease of nanobody formatting, multivalent IL23 nanobodies were assembled with properly designed linkers flanking an antihuman serum albumin nanobody, with improved hIL23 neutralization capacity in vitro and in vivo , as compared to the monovalent nanobodies. These constructs with long exposure time are excellent candidates for further developments targeting Crohn's disease, rheumatoid arthritis, and psoriasis.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques (AFMB), UMR 7257, Centre National de la Recherche Scientifique (CNRS), Marseille, France.