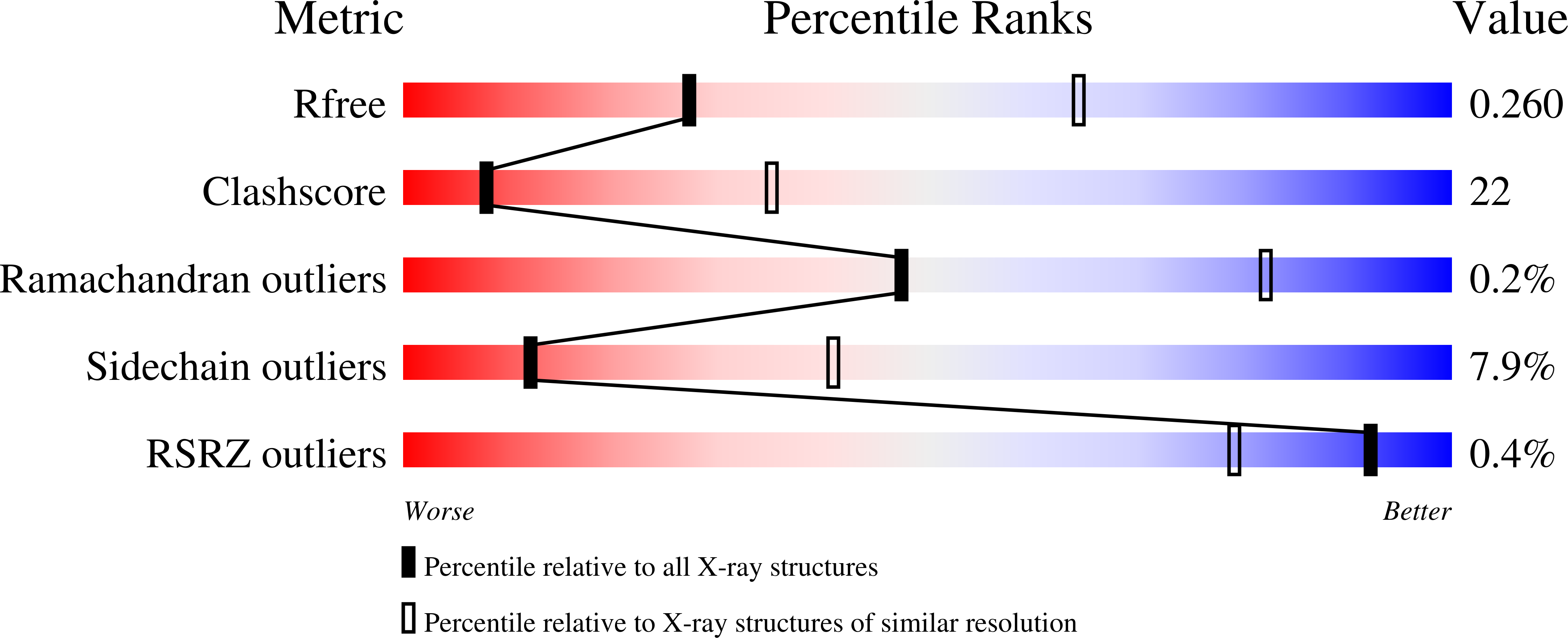
Evolution of minimal specificity and promiscuity in steroid hormone receptors.
Eick, G.N., Colucci, J.K., Harms, M.J., Ortlund, E.A., Thornton, J.W.(2012) PLoS Genet 8: e1003072-e1003072
- PubMed: 23166518
- DOI: https://doi.org/10.1371/journal.pgen.1003072
- Primary Citation of Related Structures:
4FN9, 4FNE - PubMed Abstract:
Most proteins are regulated by physical interactions with other molecules; some are highly specific, but others interact with many partners. Despite much speculation, we know little about how and why specificity/promiscuity evolves in natural proteins. It is widely assumed that specific proteins evolved from more promiscuous ancient forms and that most proteins' specificity has been tuned to an optimal state by selection. Here we use ancestral protein reconstruction to trace the evolutionary history of ligand recognition in the steroid hormone receptors (SRs), a family of hormone-regulated animal transcription factors. We resurrected the deepest ancestral proteins in the SR family and characterized the structure-activity relationships by which they distinguished among ligands. We found that that the most ancient split in SR evolution involved a discrete switch from an ancient receptor for aromatized estrogens--including xenobiotics--to a derived receptor that recognized non-aromatized progestagens and corticosteroids. The family's history, viewed in relation to the evolution of their ligands, suggests that SRs evolved according to a principle of minimal specificity: at each point in time, receptors evolved ligand recognition criteria that were just specific enough to parse the set of endogenous substances to which they were exposed. By studying the atomic structures of resurrected SR proteins, we found that their promiscuity evolved because the ancestral binding cavity was larger than the primary ligand and contained excess hydrogen bonding capacity, allowing adventitious recognition of larger molecules with additional functional groups. Our findings provide an historical explanation for the sensitivity of modern SRs to natural and synthetic ligands--including endocrine-disrupting drugs and pollutants--and show that knowledge of history can contribute to ligand prediction. They suggest that SR promiscuity may reflect the limited power of selection within real biological systems to discriminate between perfect and "good enough."
Organizational Affiliation:
Institute of Ecology and Evolution, University of Oregon, Eugene, Oregon, United States of America.