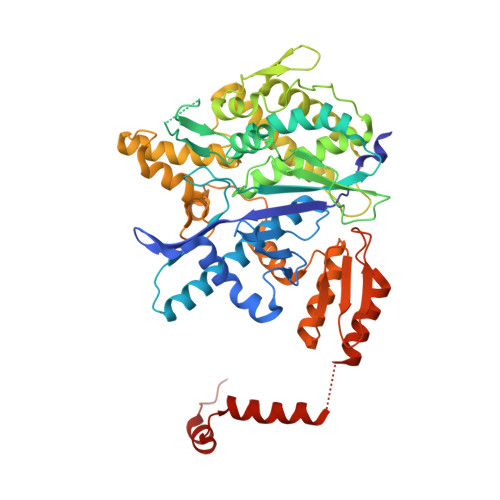
Crystal Structure of the Complex between Prokaryotic Ubiquitin-Like Protein Pup and its Ligase Pafa.
Barandun, J., Delley, C.L., Ban, N., Weber-Ban, E.(2013) J Am Chem Soc 135: 6794
- PubMed: 23601177
- DOI: https://doi.org/10.1021/ja4024012
- Primary Citation of Related Structures:
4BJR - PubMed Abstract:
Prokaryotic ubiquitin-like protein (Pup) is covalently attached to target proteins by the ligase PafA, tagging substrates for proteasomal degradation. The crystal structure of Pup in complex with PafA, reported here, reveals that a long groove wrapping around the enzyme serves as a docking site for Pup. Upon binding, the C-terminal region of the intrinsically disordered Pup becomes ordered to form two helices connected by a linker, positioning the C-terminal glutamate in the active site of PafA.
Organizational Affiliation:
Institute of Molecular Biology & Biophysics, ETH Zürich, Zürich, Switzerland.