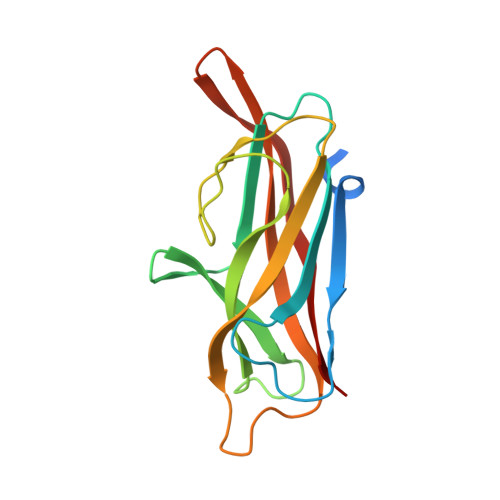
Crystal Structure of Enterotoxigenic Escherichia Coli Colonization Factor Cs6 Reveals a Novel Type of Functional Assembly.
Roy, S.P., Rahman, M.M., Yu, X.D., Tuittila, M., Knight, S.D., Zavialov, A.V.(2012) Mol Microbiol 86: 1100
- PubMed: 23046340
- DOI: https://doi.org/10.1111/mmi.12044
- Primary Citation of Related Structures:
4B9G, 4B9I, 4B9J - PubMed Abstract:
Coli surface antigen 6 (CS6) is a widely expressed enterotoxigenic Escherichia coli (ETEC) colonization factor that mediates bacterial attachment to the small intestinal epithelium. CS6 is a polymer of two protein subunits CssA and CssB, which are secreted and assembled on the cell surface via the CssC/CssD chaperone usher (CU) pathway. Here, we present an atomic resolution model for the structure of CS6 based on the results of X-ray crystallographic, spectroscopic and biochemical studies, and suggest a mechanism for CS6-mediated adhesion. We show that the CssA and CssB subunits are assembled alternately in linear fibres by the principle of donor strand complementation. This type of fibre assembly is novel for CU assembled adhesins. We also show that both subunits in the fibre bind to receptors on epithelial cells, and that CssB, but not CssA, specifically recognizes the extracellular matrix protein fibronectin. Taken together, structural and functional results suggest that CS6 is an adhesive organelle of a novel type, a hetero-polyadhesin that is capable of polyvalent attachment to different receptors.
Organizational Affiliation:
Department of Molecular Biology, Uppsala BioCenter, Swedish University of Agricultural Sciences, SE-753 24 Uppsala, Sweden.