Distinct Regions of the Pseudomonas Syringae Coiled-Coil Effector Avrrps4 are Required for Activation of Immunity.
Sohn, K.H., Hughes, R.K., Piquerez, S.J., Jones, J.D.G., Banfield, M.J.(2012) Proc Natl Acad Sci U S A 109: 16371
- PubMed: 22988101
- DOI: https://doi.org/10.1073/pnas.1212332109
- Primary Citation of Related Structures:
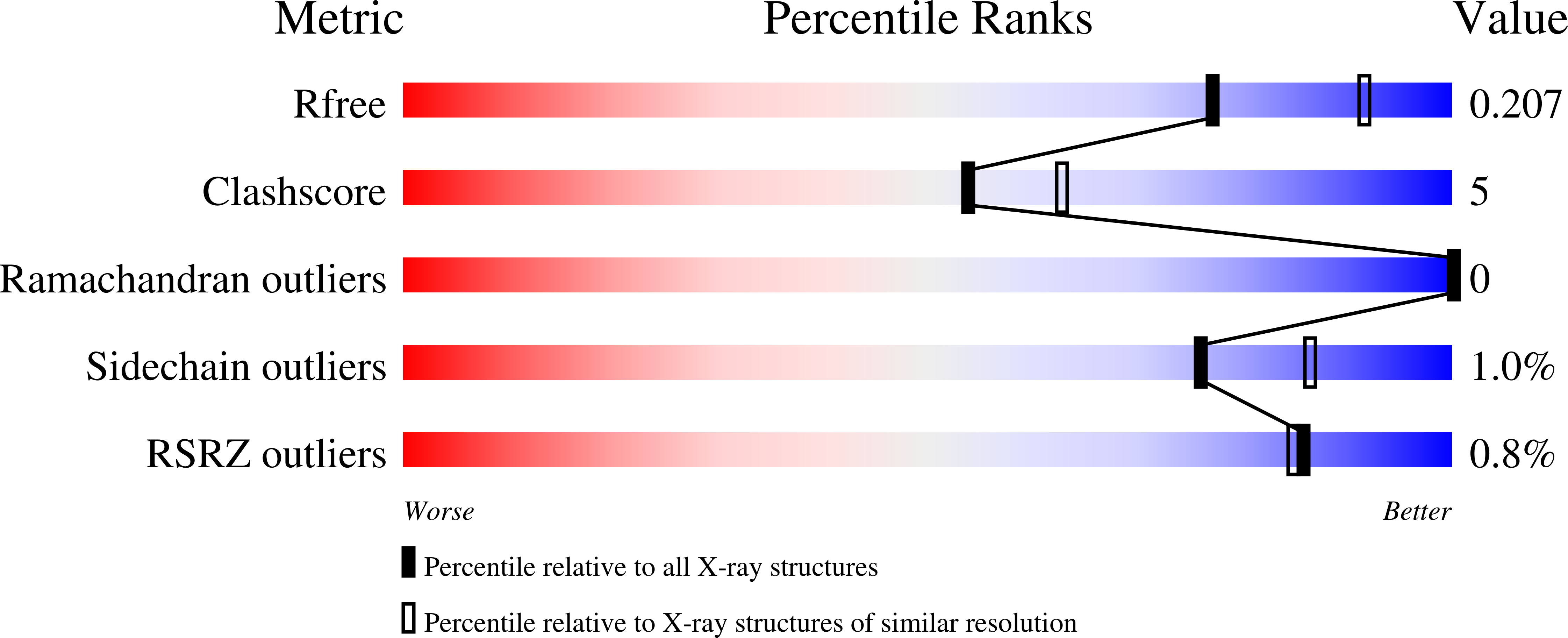
4B6X - PubMed Abstract:
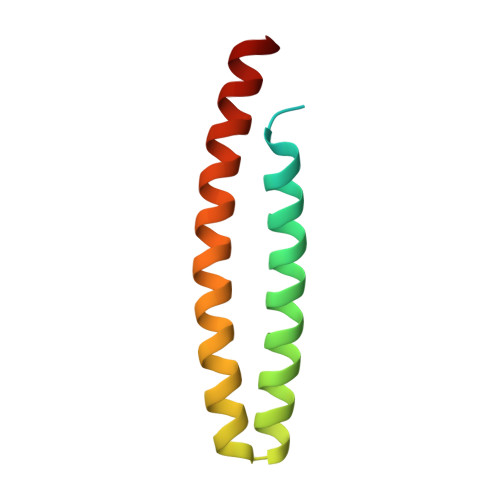
Gram-negative phytopathogenic bacteria translocate effector proteins into plant cells to subvert host defenses. These effectors can be recognized by plant nucleotide-binding-leucine-rich repeat immune receptors, triggering defense responses that restrict pathogen growth. AvrRps4, an effector protein from Pseudomonas syringae pv. pisi, triggers RPS4-dependent immunity in resistant accessions of Arabidopsis. To better understand the molecular basis of AvrRps4-triggered immunity, we determined the crystal structure of processed AvrRps4 (AvrRps4(C), residues 134-221), revealing that it forms an antiparallel α-helical coiled coil. Structure-informed mutagenesis reveals an electronegative surface patch in AvrRps4(C) required for recognition by RPS4; mutations in this region can also uncouple triggering of the hypersensitive response from disease resistance. This uncoupling may result from a lower level of defense activation, sufficient for avirulence but not for triggering a hypersensitive response. Natural variation in AvrRps4 reveals distinct recognition specificities that involve a surface-exposed residue. Recently, a direct interaction between AvrRps4 and Enhanced Disease Susceptibility 1 has been implicated in activation of immunity. However, we were unable to detect direct interaction between AvrRps4 and Enhanced Disease Susceptibility 1 after coexpression in Nicotiana benthamiana or in yeast cells. How intracellular plant immune receptors activate defense upon effector perception remains an unsolved problem. The structure of AvrRps4(C), and identification of functionally important residues for its activation of plant immunity, advances our understanding of these processes in a well-defined model pathosystem.
Organizational Affiliation:
The Sainsbury Laboratory, Norwich Research Park, Norwich NR4 7UH, United Kingdom.