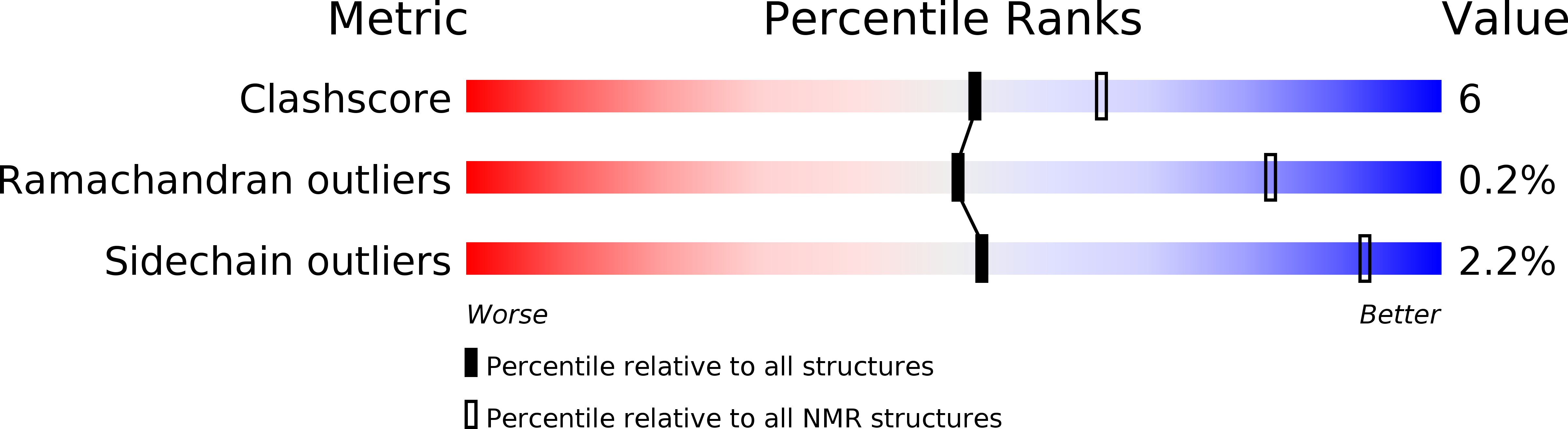Structure of the O-Glycosylated Conopeptide Cctx from Conus Consors Venom.
Hocking, H.G., Gerwig, G.J., Dutertre, S., Violette, A., Favreau, P., Stocklin, R., Kamerling, J.P., Boelens, R.(2013) Chemistry 19: 870
- PubMed: 23281027
- DOI: https://doi.org/10.1002/chem.201202713
- Primary Citation of Related Structures:
4B1Q - PubMed Abstract:
The glycopeptide CcTx, isolated from the venom of the piscivorous cone snail Conus consors, belongs to the κA-family of conopeptides. These toxins elicit excitotoxic responses in the prey by acting on voltage-gated sodium channels. The structure of CcTx, a first in the κA-family, has been determined by high-resolution NMR spectroscopy together with the analysis of its O-glycan at Ser7. A new type of glycopeptide O-glycan core structure, here registered as core type 9, containing two terminal L-galactose units {α-L-Galp-(1→4)-α-D-GlcpNAc-(1→6)-[α-L-Galp-(1→2)-β-D-Galp-(1→3)-]α-D-GalpNAc-(1→O)}, is highlighted. A sequence comparison to other putative members of the κA-family suggests that O-linked glycosylation might be more common than previously thought. This observation alone underlines the requirement for more careful and in-depth investigations into this type of post-translational modification in conotoxins.
Organizational Affiliation:
NMR Spectroscopy, Bijvoet Center for Biomolecular Research, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands. h.hocking@tum.de