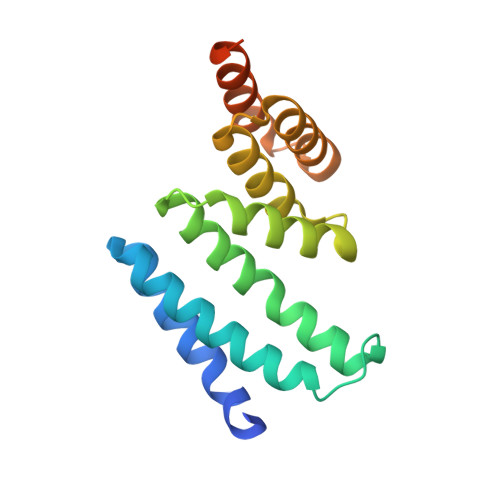
Structure of the Tpr Domain of Aip: Lack of Client Protein Interaction with the C-Terminal Alpha-7 Helix of the Tpr Domain of Aip is Sufficient for Pituitary Adenoma Predisposition.
Morgan, R.M., Hernandez-Ramirez, L.C., Trivellin, G., Zhou, L., Roe, S.M., Korbonits, M., Prodromou, C.(2012) PLoS One 7: 53339
- PubMed: 23300914
- DOI: https://doi.org/10.1371/journal.pone.0053339
- Primary Citation of Related Structures:
4AIF, 4APO - PubMed Abstract:
Mutations of the aryl hydrocarbon receptor interacting protein (AIP) have been associated with familial isolated pituitary adenomas predisposing to young-onset acromegaly and gigantism. The precise tumorigenic mechanism is not well understood as AIP interacts with a large number of independent proteins as well as three chaperone systems, HSP90, HSP70 and TOMM20. We have determined the structure of the TPR domain of AIP at high resolution, which has allowed a detailed analysis of how disease-associated mutations impact on the structural integrity of the TPR domain. A subset of C-terminal α-7 helix (Cα-7h) mutations, R304* (nonsense mutation), R304Q, Q307* and R325Q, a known site for AhR and PDE4A5 client-protein interaction, occur beyond those that interact with the conserved MEEVD and EDDVE sequences of HSP90 and TOMM20. These C-terminal AIP mutations appear to only disrupt client-protein binding to the Cα-7h, while chaperone binding remains unaffected, suggesting that failure of client-protein interaction with the Cα-7h is sufficient to predispose to pituitary adenoma. We have also identified a molecular switch in the AIP TPR-domain that allows recognition of both the conserved HSP90 motif, MEEVD, and the equivalent sequence (EDDVE) of TOMM20.
Organizational Affiliation:
Genome Damage and Stability Centre, University of Sussex, Brighton, United Kingdom.