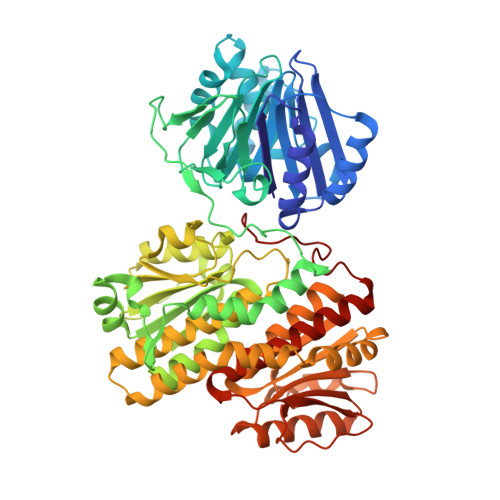
Glutamine Binding Opens the Ammonia Channel and Activates Glucosamine-6P Synthase
Mouilleron, S., Badet-Denisot, M.A., Golinelli-Pimpaneau, B.(2006) J Biol Chem 281: 4404
- PubMed: 16339762
- DOI: https://doi.org/10.1074/jbc.M511689200
- Primary Citation of Related Structures:
2J6H, 4AMV - PubMed Abstract:
Glucosamine-6P synthase catalyzes the synthesis of glucosamine-6P from fructose-6P and glutamine and uses a channel to transfer ammonia from its glutaminase to its synthase active site. X-ray structures of glucosamine-6P synthase have been determined at 2.05 Angstroms resolution in the presence of fructose-6P and at 2.35 Angstroms resolution in the presence of fructose-6P and 6-diazo-5-oxo-L-norleucine, a glutamine affinity analog that covalently modifies the N-terminal catalytic cysteine, therefore mimicking the gamma-glutamyl-thioester intermediate formed during hydrolysis of glutamine. The fixation of the glutamine analog activates the enzyme through several major structural changes: 1) the closure of a loop to shield the glutaminase site accompanied by significant domain hinging, 2) the activation of catalytic residues involved in glutamine hydrolysis, i.e. the alpha-amino group of Cys-1 and Asn-98 that is positioned to form the oxyanion hole, and 3) a 75 degrees rotation of the Trp-74 indole group that opens the ammonia channel.
Organizational Affiliation:
Laboratoire d'Enzymologie et de Biochimie Structurales, Unite Propre de Recherche 9063, France.