Structures and Binding Specificity of Galactose- and Mannose-Binding Lectins from Champedak: Differences from Jackfruit Lectins
Gabrielsen, M., Abdul-Rahman, P.S., Othman, S., Hashim, O.H., Cogdell, R.J.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 709
- PubMed: 24915077
- DOI: https://doi.org/10.1107/S2053230X14008966
- Primary Citation of Related Structures:
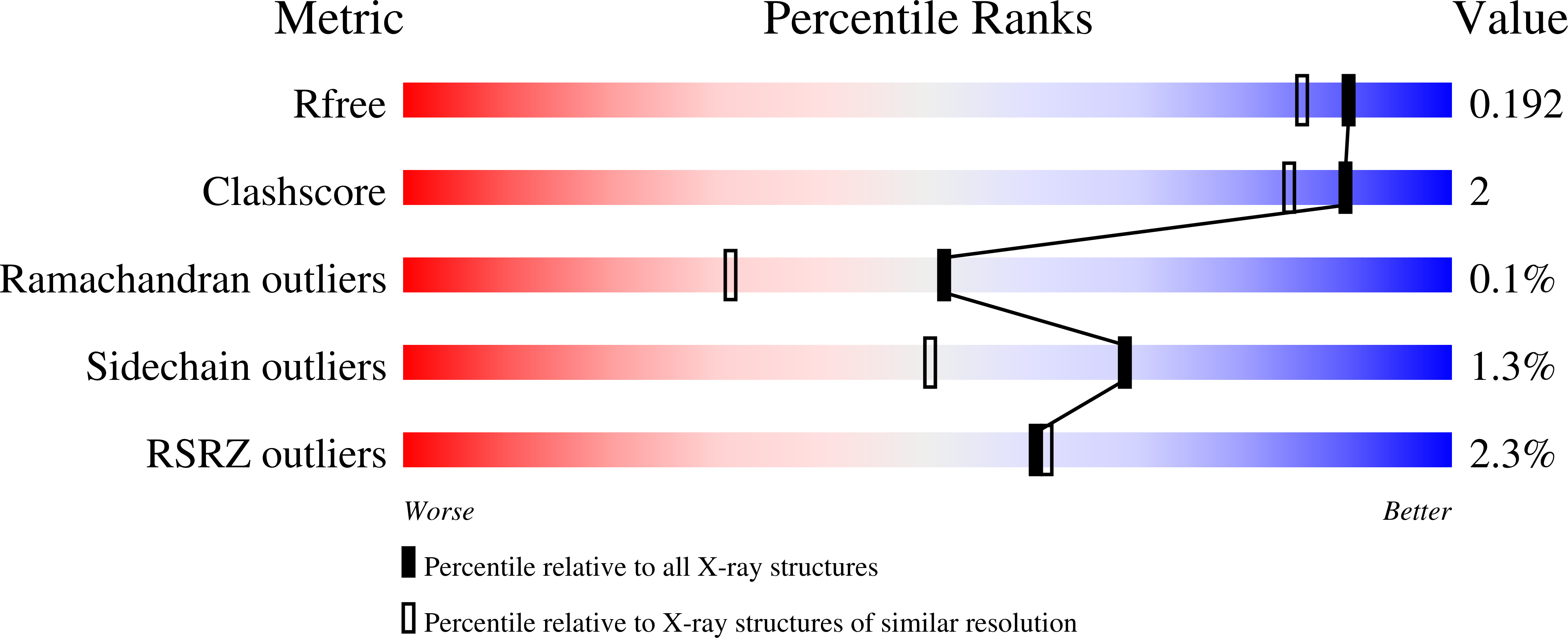
4AK4, 4AKB, 4AKC, 4AKD - PubMed Abstract:
Galactose-binding and mannose-binding lectins from the champedak fruit, which is native to South-east Asia, exhibit useful potential clinical applications. The specificity of the two lectins for their respective ligands allows the detection of potential cancer biomarkers and monitoring of the glycosylated state of proteins in human serum and/or urine. To fully understand and expand the use of these natural proteins, their complete sequences and crystal structures are presented here, together with details of sugar binding.
Organizational Affiliation:
Institute of Molecular, Cell and Systems Biology, University of Glasgow, 120 University Avenue, Glasgow G12 8TA, Scotland.