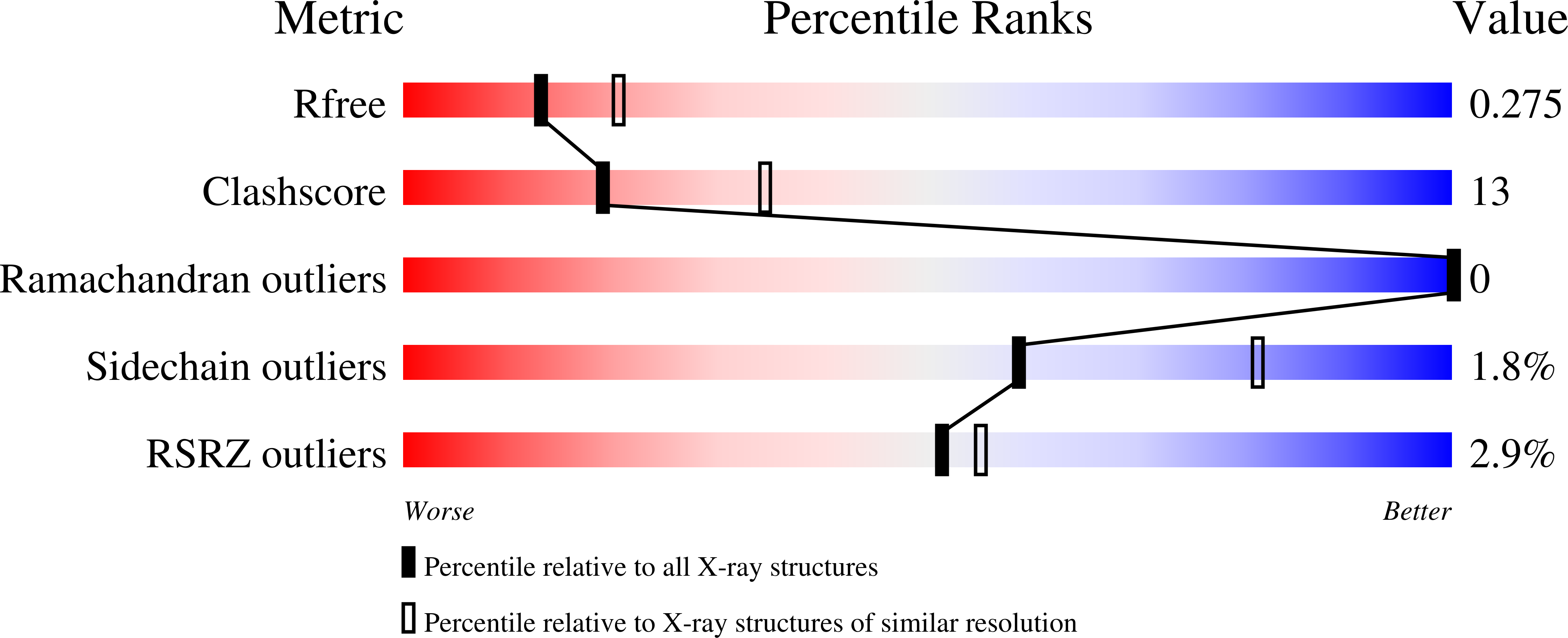
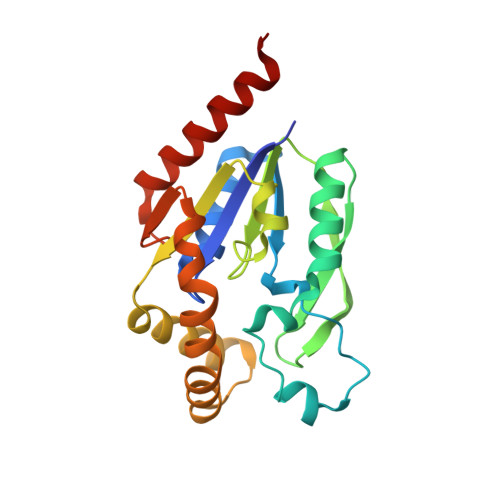
Indispensable residue for uridine binding in the uridine-cytidine kinase family.
Tomoike, F., Nakagawa, N., Fukui, K., Yano, T., Kuramitsu, S., Masui, R.(2017) Biochem Biophys Rep 11: 93-98
- PubMed: 28955773
- DOI: https://doi.org/10.1016/j.bbrep.2017.07.002
- Primary Citation of Related Structures:
3W8R - PubMed Abstract:
Uridine-cytidine kinase (UCK), including human UCK2, are a family of enzymes that generally phosphorylate both uridine and cytidine. However, UCK of Thermus thermophilus HB8 (ttCK) phosphorylates only cytidine. This cytidine-restricted activity is thought to depend on Tyr93, although the precise mechanism remains unresolved. Exhaustive mutagenesis of Tyr93 in ttCK revealed that the uridine phosphorylation activity was restored only by replacement of Tyr93 with His or Gln. Replacement of His117 in human UCK2, corresponding to residue Tyr93 in ttCK, by Tyr resulted in a loss of uridine phosphorylation activity. These findings indicated that uridine phosphorylation activity commonly depends on a single residue in the UCK family.
Organizational Affiliation:
Graduate School of Frontier Biosciences, Osaka University, 1-3 Yamadaoka, Suita, Osaka 565-0871, Japan.