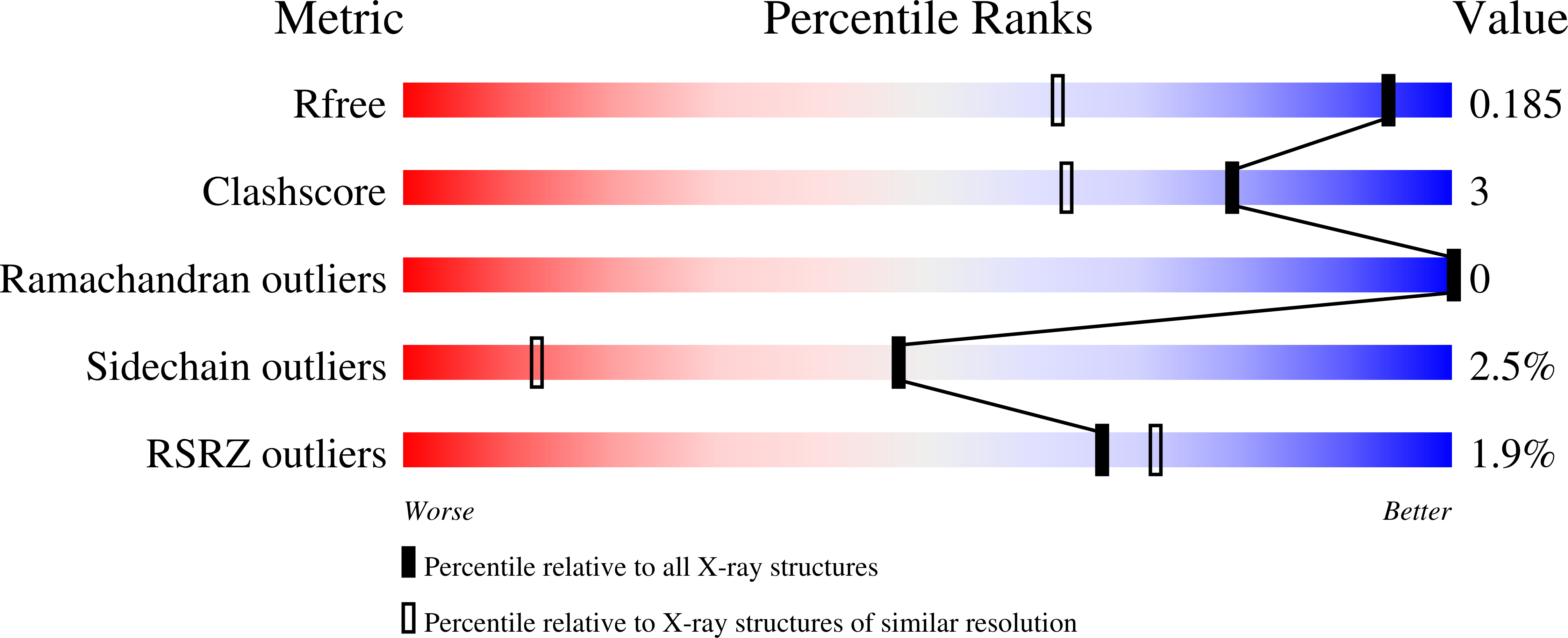
Carbonyl Sulfide Hydrolase from Thiobacillus thioparus Strain THI115 Is One of the beta-Carbonic Anhydrase Family Enzymes
Ogawa, T., Noguchi, K., Saito, M., Nagahata, Y., Kato, H., Ohtaki, A., Nakayama, H., Dohmae, N., Matsushita, Y., Odaka, M., Yohda, M., Nyunoya, H., Katayama, Y.(2013) J Am Chem Soc 135: 3818-3825
- PubMed: 23406161
- DOI: https://doi.org/10.1021/ja307735e
- Primary Citation of Related Structures:
3VQJ, 3VRK - PubMed Abstract:
Carbonyl sulfide (COS) is an atmospheric trace gas leading to sulfate aerosol formation, thereby participating in the global radiation balance and ozone chemistry, but its biological sinks are not well understood. Thiobacillus thioparus strain THI115 can grow on thiocyanate (SCN(-)) as its sole energy source. Previously, we showed that SCN(-) is first converted to COS by thiocyanate hydrolase in T. thioparus strain THI115. In the present work, we purified, characterized, and determined the crystal structure of carbonyl sulfide hydrolase (COSase), which is responsible for the degradation of COS to H2S and CO2, the second step of SCN(-) assimilation. COSase is a homotetramer composed of a 23.4 kDa subunit containing a zinc ion in its catalytic site. The amino acid sequence of COSase is homologous to the β-class carbonic anhydrases (β-CAs). Although the crystal structure including the catalytic site resembles those of the β-CAs, CO2 hydration activity of COSase is negligible compared to those of the β-CAs. The α5 helix and the extra loop (Gly150-Pro158) near the N-terminus of the α6 helix narrow the substrate pathway, which could be responsible for the substrate specificity. The k(cat)/K(m) value, 9.6 × 10(5) s(-1) M(-1), is comparable to those of the β-CAs. COSase hydrolyzes COS over a wide concentration range, including the ambient level, in vitro and in vivo. COSase and its structurally related enzymes are distributed in the clade D in the phylogenetic tree of β-CAs, suggesting that COSase and its related enzymes are one of the catalysts responsible for the global sink of COS.
Organizational Affiliation:
Department of Science of Resources and Environment, United Graduate School of Agricultural Science, Tokyo University of Agriculture and Technology, Fuchu, Tokyo 183-8509, Japan.