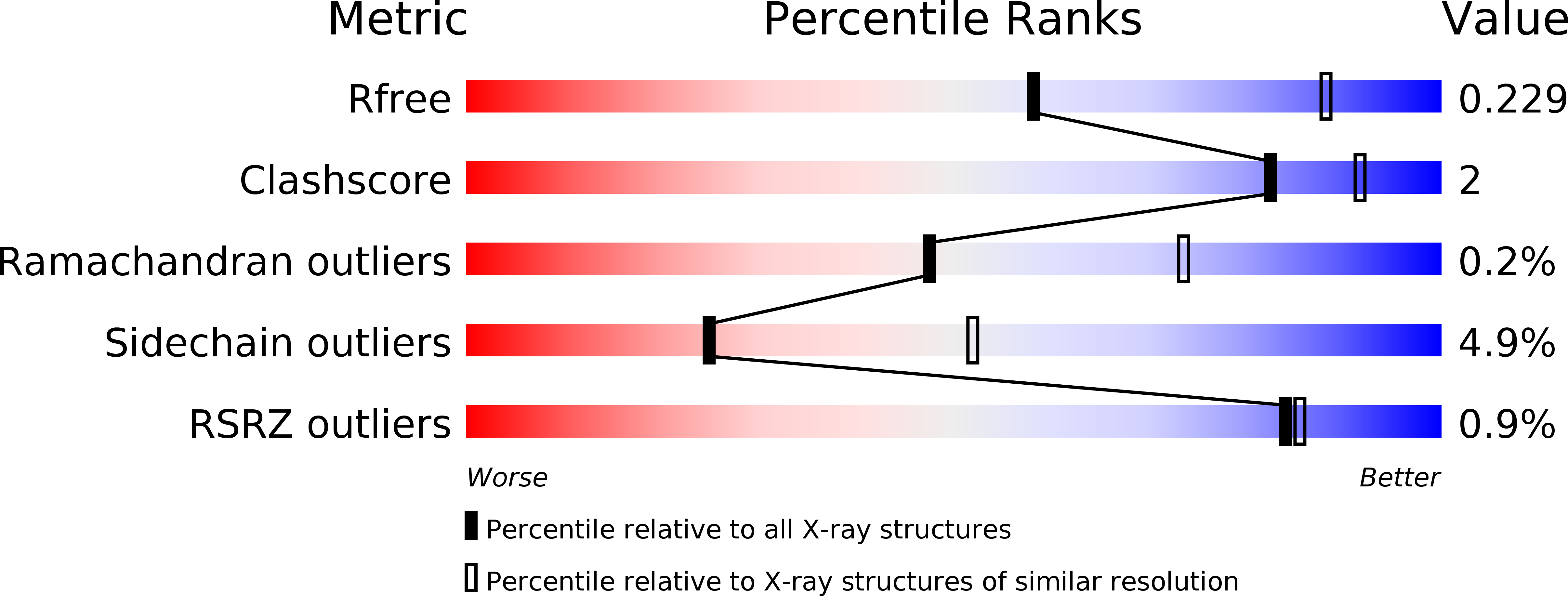
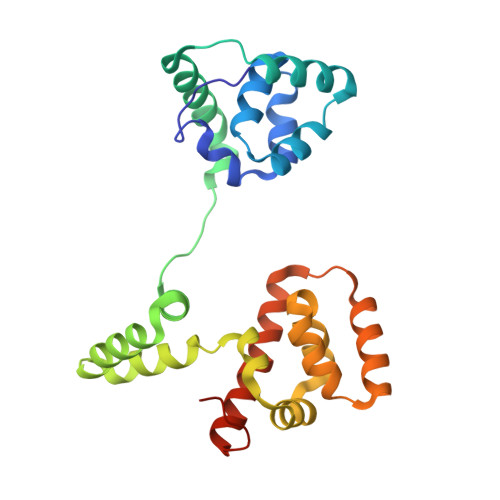
Multiple conformations of the FliG C-terminal domain provide insight into flagellar motor switching
Lam, K.H., Ip, W.S., Lam, Y.W., Chan, S.O., Ling, T.K.W., Au, S.W.N.(2012) Structure 20: 315-325
- PubMed: 22325779
- DOI: https://doi.org/10.1016/j.str.2011.11.020
- Primary Citation of Related Structures:
3USW, 3USY - PubMed Abstract:
Bacterial flagellar switching between counterclockwise and clockwise directions is mediated by the coupling of the chemotactic system and the motor switch complex. The conformational changes of FliG are closely associated with this switching mechanism. We present two crystal structures of FliG(MC) from Helicobacter pylori, each showing distinct domain orientations from previously solved structures. A 180° rotation of the charged ridge-containing C-terminal subdomain FliG(Cα1-6) that is prompted by the rotational freedom of Met245 psi and Phe246 phi at the MFXF motif was revealed. Studies on the swarming and swimming behavior of Escherichia coli mutants further identified the importance of the ₂₄₅MFXF₂₄₈ motif and a highly conserved residue, Asn216, in motor switching. Additionally, multiple conformations of FliG(Cα1-6) were demonstrated by intramolecular cysteine crosslinking. The conformational flexibility of FliGc leads us to propose a model that accounts for the symmetrical torque generation process and for the dynamics of the motor.
Organizational Affiliation:
Centre for Protein Science and Crystallography, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong.