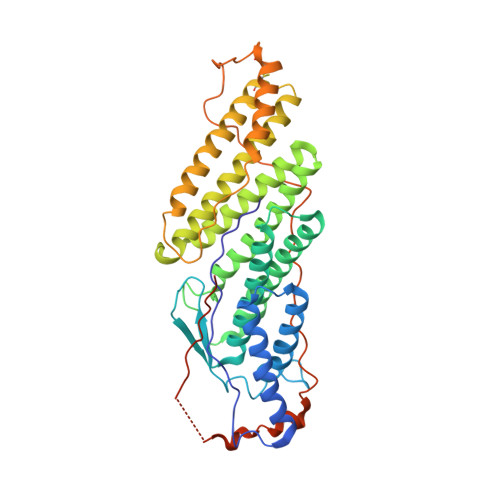
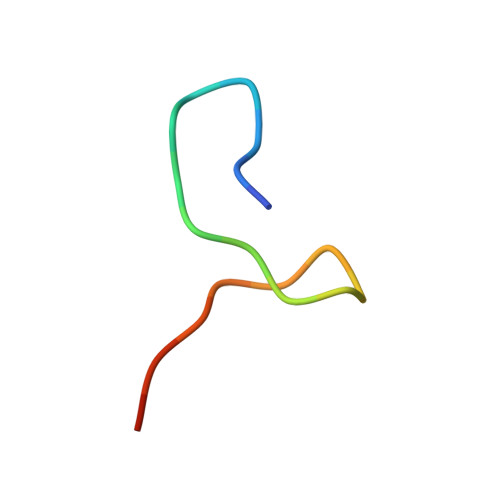
Two Distinct Binding Modes Define the Interaction of Brox with the C-Terminal Tails of CHMP5 and CHMP4B.
Mu, R., Dussupt, V., Jiang, J., Sette, P., Rudd, V., Chuenchor, W., Bello, N.F., Bouamr, F., Xiao, T.S.(2012) Structure 20: 887-898
- PubMed: 22484091
- DOI: https://doi.org/10.1016/j.str.2012.03.001
- Primary Citation of Related Structures:
3ULY, 3UM0, 3UM1, 3UM2, 3UM3 - PubMed Abstract:
Interactions of the CHMP protein carboxyl terminal tails with effector proteins play important roles in retroviral budding, cytokinesis, and multivesicular body biogenesis. Here we demonstrate that hydrophobic residues at the CHMP4B C-terminal amphipathic α helix bind a concave surface of Brox, a mammalian paralog of Alix. Unexpectedly, CHMP5 was also found to bind Brox and specifically recruit endogenous Brox to detergent-resistant membrane fractions through its C-terminal 20 residues. Instead of an α helix, the CHMP5 C-terminal tail adopts a tandem β-hairpin structure that binds Brox at the same site as CHMP4B. Additional Brox:CHMP5 interface is furnished by a unique CHMP5 hydrophobic pocket engaging the Brox residue Y348 that is not conserved among the Bro1 domains. Our studies thus unveil a β-hairpin conformation of the CHMP5 protein C-terminal tail, and provide insights into the overlapping but distinct binding profiles of ESCRT-III and the Bro1 domain proteins.
Organizational Affiliation:
Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, MD 20892, USA.