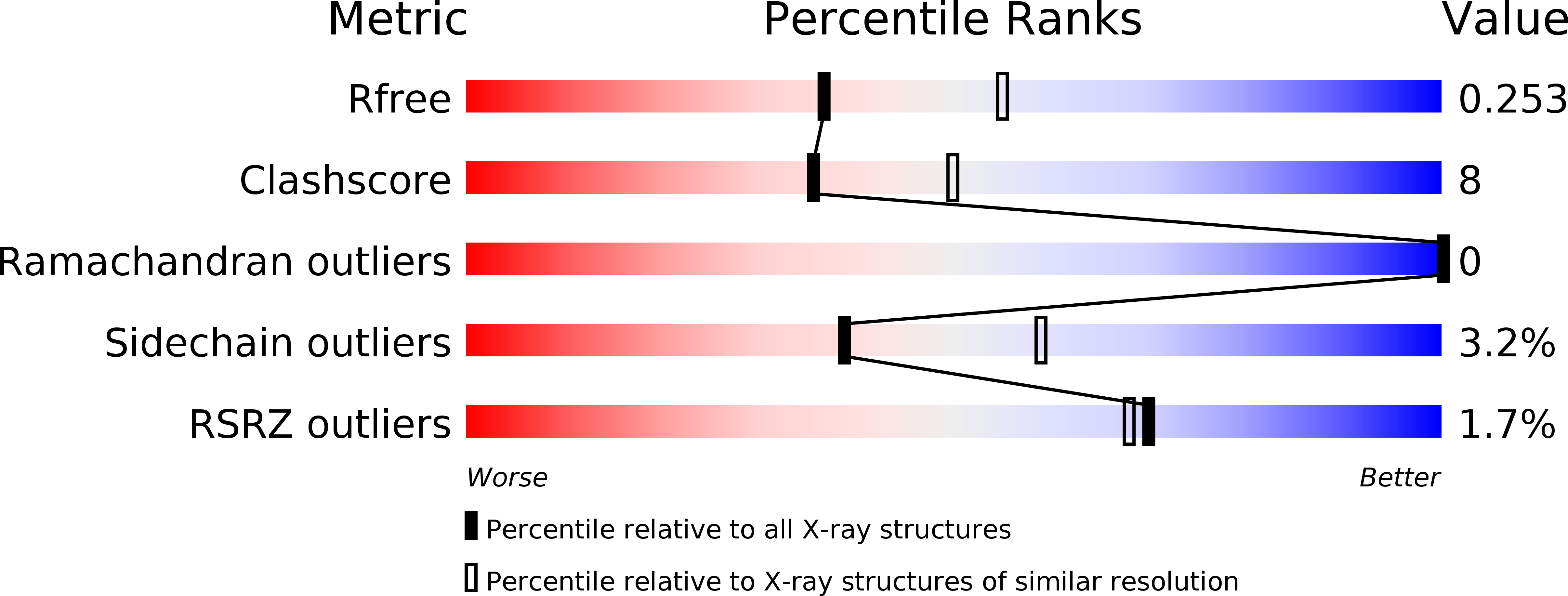
A novel "open-form" structure of sortaseC from Streptococcus suis.
Lu, G., Qi, J., Gao, F., Yan, J., Tang, J., Gao, G.F.(2011) Proteins 79: 2764-2769
- PubMed: 21721048
- DOI: https://doi.org/10.1002/prot.23093
- Primary Citation of Related Structures:
3RE9
Organizational Affiliation:
CAS Key Laboratory of Pathogenic Microbiology and Immunology (CASPMI), Institute of Microbiology, Chinese Academy of Sciences (CAS), Beijing 100101, China.