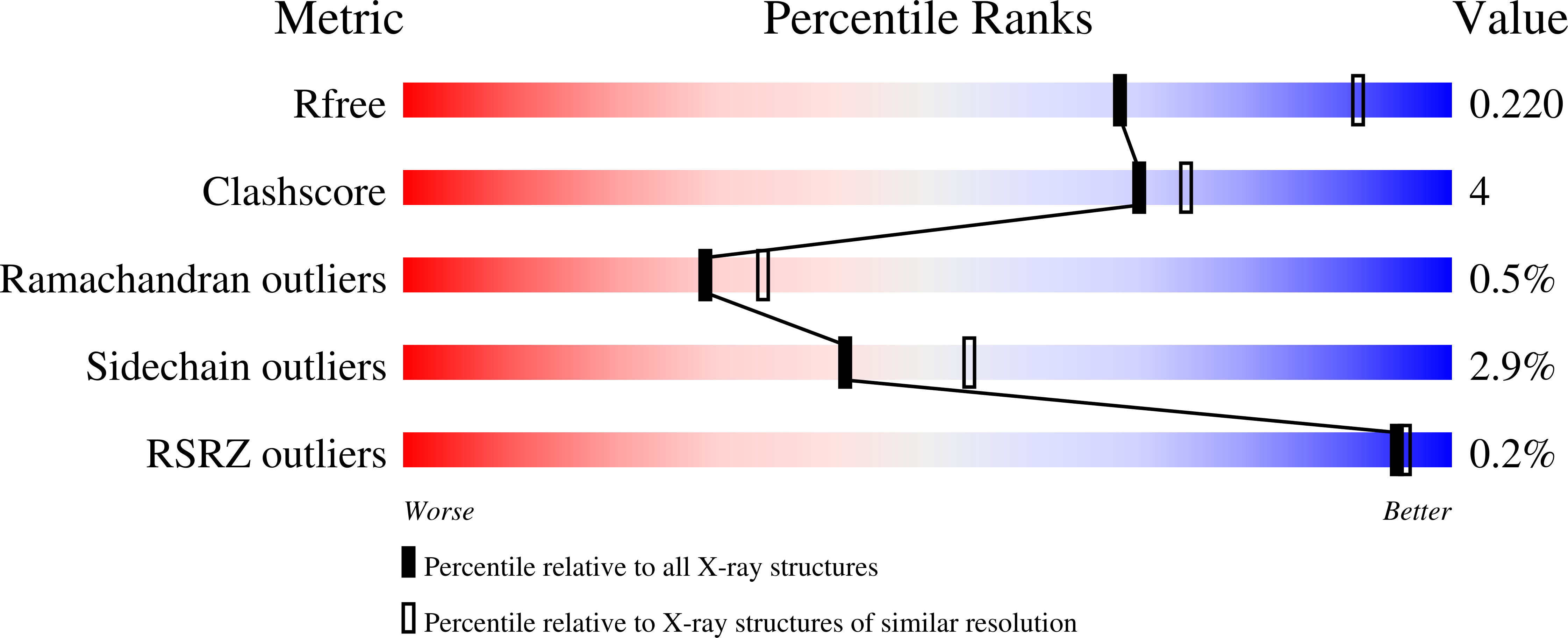
Structure of the Lassa virus nucleoprotein revealed by X-ray crystallography, small-angle X-ray scattering, and electron microscopy.
Brunotte, L., Kerber, R., Shang, W., Hauer, F., Hass, M., Gabriel, M., Lelke, M., Busch, C., Stark, H., Svergun, D.I., Betzel, C., Perbandt, M., Gunther, S.(2011) J Biol Chem 286: 38748-38756
- PubMed: 21917929
- DOI: https://doi.org/10.1074/jbc.M111.278838
- Primary Citation of Related Structures:
3R3L - PubMed Abstract:
The nucleoprotein (NP) of Lassa virus (LASV) strain AV was expressed in a recombinant baculovirus system. The crystal structure of full-length NP was solved at a resolution of 2.45 Å. The overall fold corresponds to that of NP of LASV strain Josiah (Qi, X., Lan, S., Wang, W., Schelde, L. M., Dong, H., Wallat, G. D., Ly, H., Liang, Y., and Dong, C. (2010) Nature 468, 779-783) with a root mean square deviation of 0.67 Å for all atoms (6.3% difference in primary sequence). As the packing in the crystal offers two different trimer architectures for the biological assembly, the quaternary structure of NP in solution was determined by small-angle x-ray scattering and EM. After classification and averaging of >6000 EM raw images, trimeric centrosymmetric structures were obtained, which correspond in size and shape to one trimer in the crystal structure formed around a crystallographic 3-fold rotation axis (symmetric trimer). The symmetric trimer is also a good model for the small-angle x-ray scattering data and could be well embedded into the ab initio model. The N-terminal domain of NP contains a deep nucleotide-binding cavity that has been proposed to bind cellular cap structures for priming viral mRNA synthesis. All residues implicated in m(7)GpppN binding were exchanged, and the transcription/replication phenotype of the NP mutant was tested using a LASV replicon system. None of the mutants showed a specific defect in mRNA expression; most were globally defective in RNA synthesis. In conclusion, we describe the full-length crystal structure and the quaternary structure in solution of LASV NP. The nucleotide-binding pocket of NP could not be assigned a specific role in viral mRNA synthesis.
Organizational Affiliation:
Department of Virology, Bernhard Nocht Institute for Tropical Medicine, 20359 Hamburg, Germany.