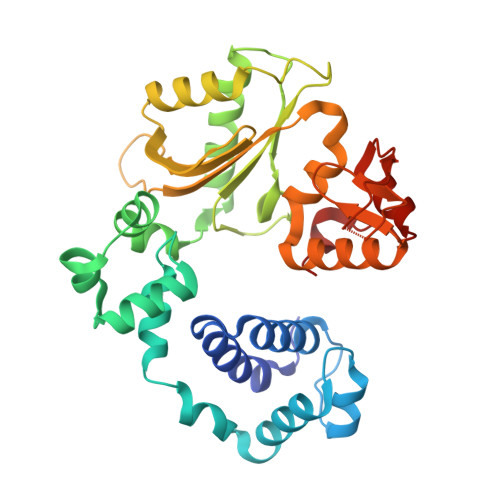
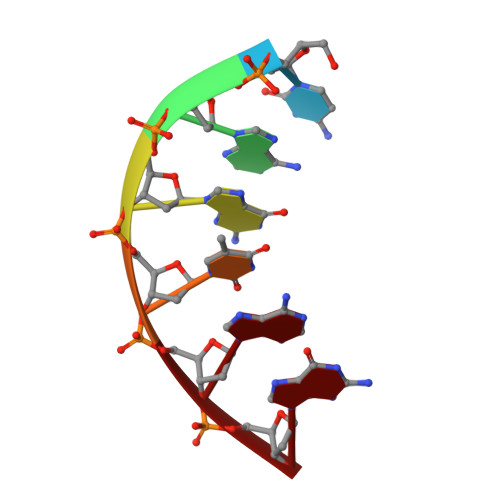
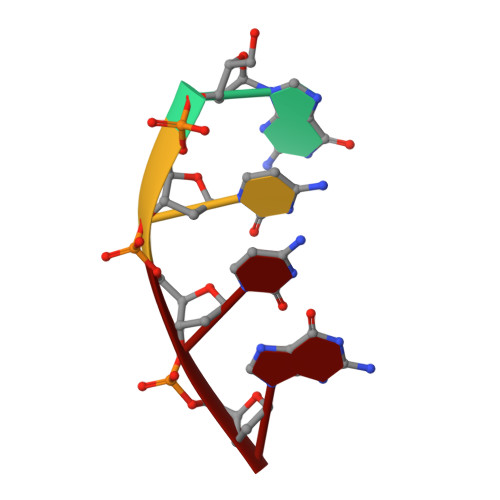
Replication infidelity via a mismatch with Watson-Crick geometry.
Bebenek, K., Pedersen, L.C., Kunkel, T.A.(2011) Proc Natl Acad Sci U S A 108: 1862-1867
- PubMed: 21233421
- DOI: https://doi.org/10.1073/pnas.1012825108
- Primary Citation of Related Structures:
3PML, 3PMN, 3PNC - PubMed Abstract:
In describing the DNA double helix, Watson and Crick suggested that "spontaneous mutation may be due to a base occasionally occurring in one of its less likely tautomeric forms." Indeed, among many mispairing possibilities, either tautomerization or ionization of bases might allow a DNA polymerase to insert a mismatch with correct Watson-Crick geometry. However, despite substantial progress in understanding the structural basis of error prevention during polymerization, no DNA polymerase has yet been shown to form a natural base-base mismatch with Watson-Crick-like geometry. Here we provide such evidence, in the form of a crystal structure of a human DNA polymerase λ variant poised to misinsert dGTP opposite a template T. All atoms needed for catalysis are present at the active site and in positions that overlay with those for a correct base pair. The mismatch has Watson-Crick geometry consistent with a tautomeric or ionized base pair, with the pH dependence of misinsertion consistent with the latter. The results support the original idea that a base substitution can originate from a mismatch having Watson-Crick geometry, and they suggest a common catalytic mechanism for inserting a correct and an incorrect nucleotide. A second structure indicates that after misinsertion, the now primer-terminal G • T mismatch is also poised for catalysis but in the wobble conformation seen in other studies, indicating the dynamic nature of the pathway required to create a mismatch in fully duplex DNA.
Organizational Affiliation:
Laboratory of Molecular Genetics, Department of Health and Human Services, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.