Crystal structures of active and inhibitor-bound human Casp6
Mueller, I., Lamers, M.B.A.C., Ritchie, A.J., Dominquez, C., Munoz, I., Maillard, M., Kiselyov, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| caspase-6 | 179 | Homo sapiens | Mutation(s): 0 Gene Names: CASP6 | 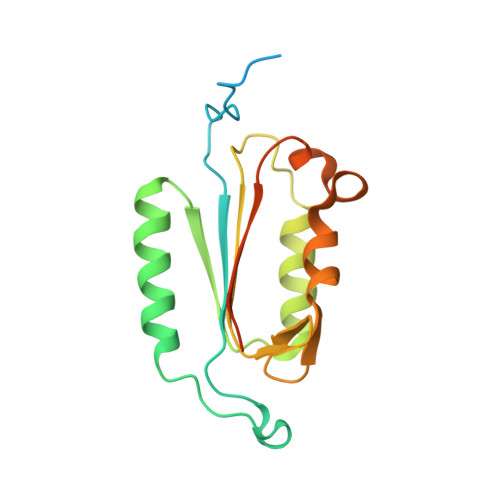 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P55212 (Homo sapiens) Explore P55212 Go to UniProtKB: P55212 | |||||
PHAROS: P55212 GTEx: ENSG00000138794 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P55212 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| caspase-6 | 108 | Homo sapiens | Mutation(s): 0 Gene Names: CASP6 | 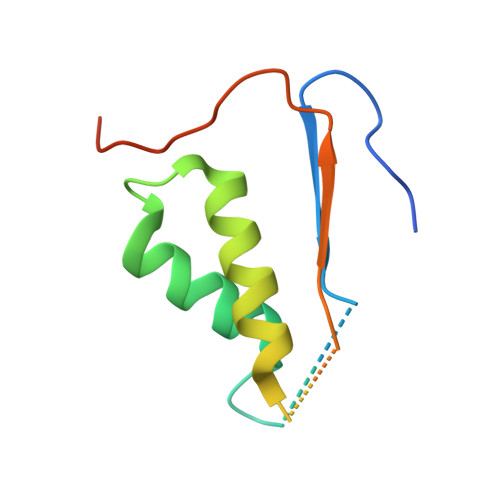 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P55212 (Homo sapiens) Explore P55212 Go to UniProtKB: P55212 | |||||
PHAROS: P55212 GTEx: ENSG00000138794 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P55212 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 81.23 | α = 90 |
| b = 161.24 | β = 94.8 |
| c = 88.92 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| CrystalClear | data collection |