Structure-function analysis of the C-terminal domain of CNM67, a core component of the Saccharomyces cerevisiae spindle pole body.
Klenchin, V.A., Frye, J.J., Jones, M.H., Winey, M., Rayment, I.(2011) J Biol Chem 286: 18240-18250
- PubMed: 21454609
- DOI: https://doi.org/10.1074/jbc.M111.227371
- Primary Citation of Related Structures:
3OA7 - PubMed Abstract:
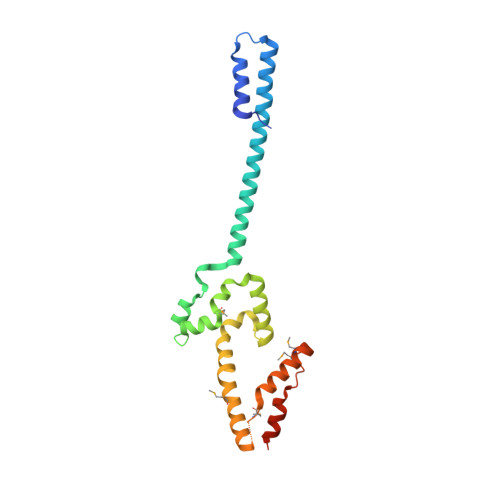
The spindle pole body of the budding yeast Saccharomyces cerevisiae has served as a model system for understanding microtubule organizing centers, yet very little is known about the molecular structure of its components. We report here the structure of the C-terminal domain of the core component Cnm67 at 2.3 Å resolution. The structure determination was aided by a novel approach to crystallization of proteins containing coiled-coils that utilizes globular domains to stabilize the coiled-coils. This enhances their solubility in Escherichia coli and improves their crystallization. The Cnm67 C-terminal domain (residues Asn-429-Lys-581) exhibits a previously unseen dimeric, interdigitated, all α-helical fold. In vivo studies demonstrate that this domain alone is able to localize to the spindle pole body. In addition, the structure reveals a large functionally indispensable positively charged surface patch that is implicated in spindle pole body localization. Finally, the C-terminal eight residues are disordered but are critical for protein folding and structural stability.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin, Madison, Wisconsin 53706, USA.