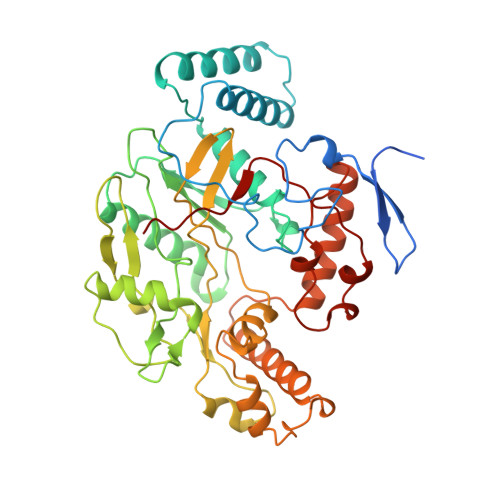
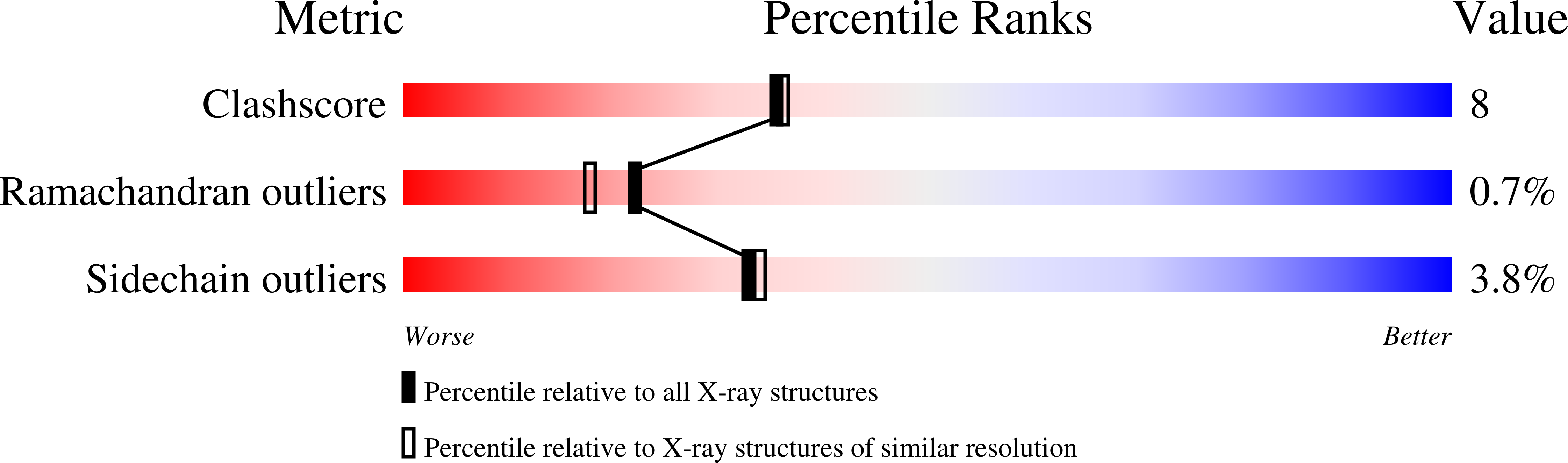Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
Raman, C.S., Li, H., Martasek, P., Kral, V., Masters, B.S., Poulos, T.L.(1998) Cell 95: 939-950
- PubMed: 9875848
- DOI: https://doi.org/10.1016/s0092-8674(00)81718-3
- Primary Citation of Related Structures:
1NSE, 2NSE, 3NSE, 4NSE - PubMed Abstract:
Nitric oxide, a key signaling molecule, is produced by a family of enzymes collectively called nitric oxide synthases (NOS). Here, we report the crystal structure of the heme domain of endothelial NOS in tetrahydrobiopterin (H4B)-free and -bound forms at 1.95 A and 1.9 A resolution, respectively. In both structures a zinc ion is tetrahedrally coordinated to pairs of symmetry-related cysteine residues at the dimer interface. The phylogenetically conserved Cys-(X)4-Cys motif and its strategic location establish a structural role for the metal center in maintaining the integrity of the H4B-binding site. The unexpected recognition of the substrate, L-arginine, at the H4B site indicates that this site is poised to stabilize a positively charged pterin ring and suggests a model involving a cationic pterin radical in the catalytic cycle.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine 92697-3900, USA.