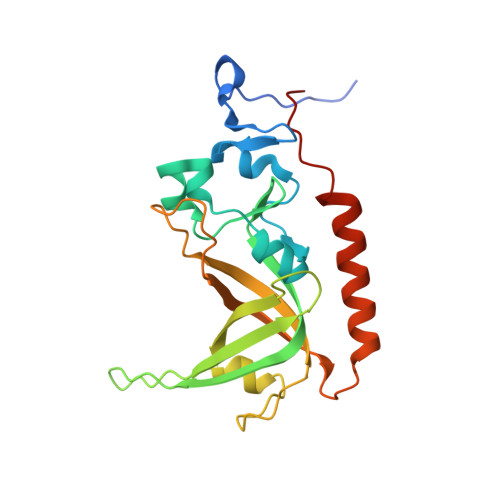
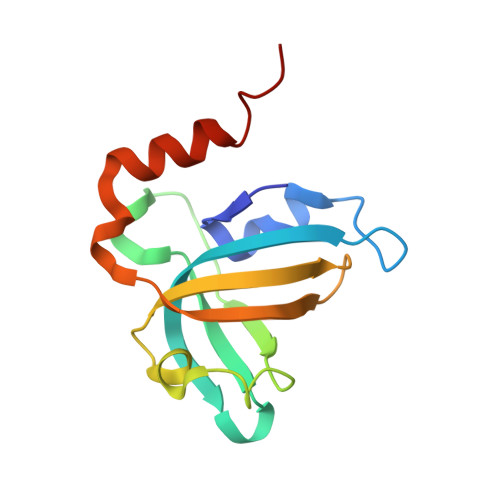
Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres.
Sun, J., Yu, E.Y., Yang, Y., Confer, L.A., Sun, S.H., Wan, K., Lue, N.F., Lei, M.(2009) Genes Dev 23: 2900-2914
- PubMed: 20008938
- DOI: https://doi.org/10.1101/gad.1851909
- Primary Citation of Related Structures:
3KEY, 3KF6, 3KF8 - PubMed Abstract:
In budding yeast, Cdc13, Stn1, and Ten1 form a heterotrimeric complex (CST) that is essential for telomere protection and maintenance. Previous bioinformatics analysis revealed a putative oligonucleotide/oligosaccharide-binding (OB) fold at the N terminus of Stn1 (Stn1N) that shows limited sequence similarity to the OB fold of Rpa2, a subunit of the eukaryotic ssDNA-binding protein complex replication protein A (RPA). Here we present functional and structural analyses of Stn1 and Ten1 from multiple budding and fission yeast. The crystal structure of the Candida tropicalis Stn1N complexed with Ten1 demonstrates an Rpa2N-Rpa3-like complex. In both structures, the OB folds of the two components pack against each other through interactions between two C-terminal helices. The structure of the C-terminal domain of Saccharomyces cerevisiae Stn1 (Stn1C) was found to comprise two related winged helix-turn-helix (WH) motifs, one of which is most similar to the WH motif at the C terminus of Rpa2, again supporting the notion that Stn1 resembles Rpa2. The crystal structure of the fission yeast Schizosaccharomyces pombe Stn1N-Ten1 complex exhibits a virtually identical architecture as the C. tropicalis Stn1N-Ten1. Functional analyses of the Candida albicans Stn1 and Ten1 proteins revealed critical roles for these proteins in suppressing aberrant telomerase and recombination activities at telomeres. Mutations that disrupt the Stn1-Ten1 interaction induce telomere uncapping and abolish the telomere localization of Ten1. Collectively, our structural and functional studies illustrate that, instead of being confined to budding yeast telomeres, the CST complex may represent an evolutionarily conserved RPA-like telomeric complex at the 3' overhangs that works in parallel with or instead of the well-characterized POT1-TPP1/TEBPalpha-beta complex.
Organizational Affiliation:
Howard Hughes Medical Institute, University of Michigan Medical School, Ann Arbor, Michigan 48109, USA.