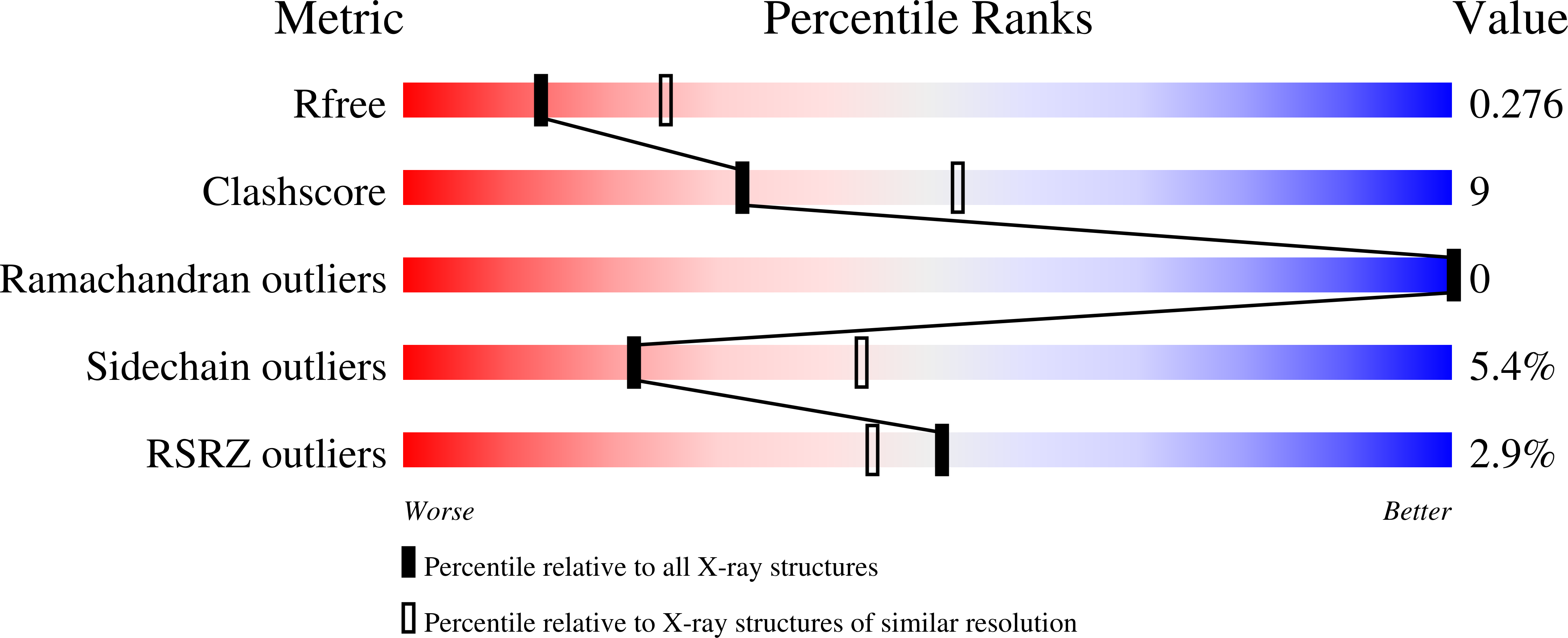
The crystal structure of the RA domain of FLJ10324 (RADIL)
Wisniewska, M., Lehtio, L., Andersson, J., Arrowsmith, C.H., Collins, R., Dahlgren, L.G., Edwards, A.M., Flodin, S., Flores, A., Graslund, S., Hammarstrom, M., Johansson, A., Johansson, I., Karlberg, T., Kotenyova, T., Moche, M., Nilsson, M.E., Nordlund, P., Nyman, T., Olesen, K., Persson, C., Sagemark, J., Schueler, H., Thorsell, A.G., Tresaugues, L., van den Berg, S., Weigelt, J., Welin, M., Wikstrom, M., Berglund, H.To be published.