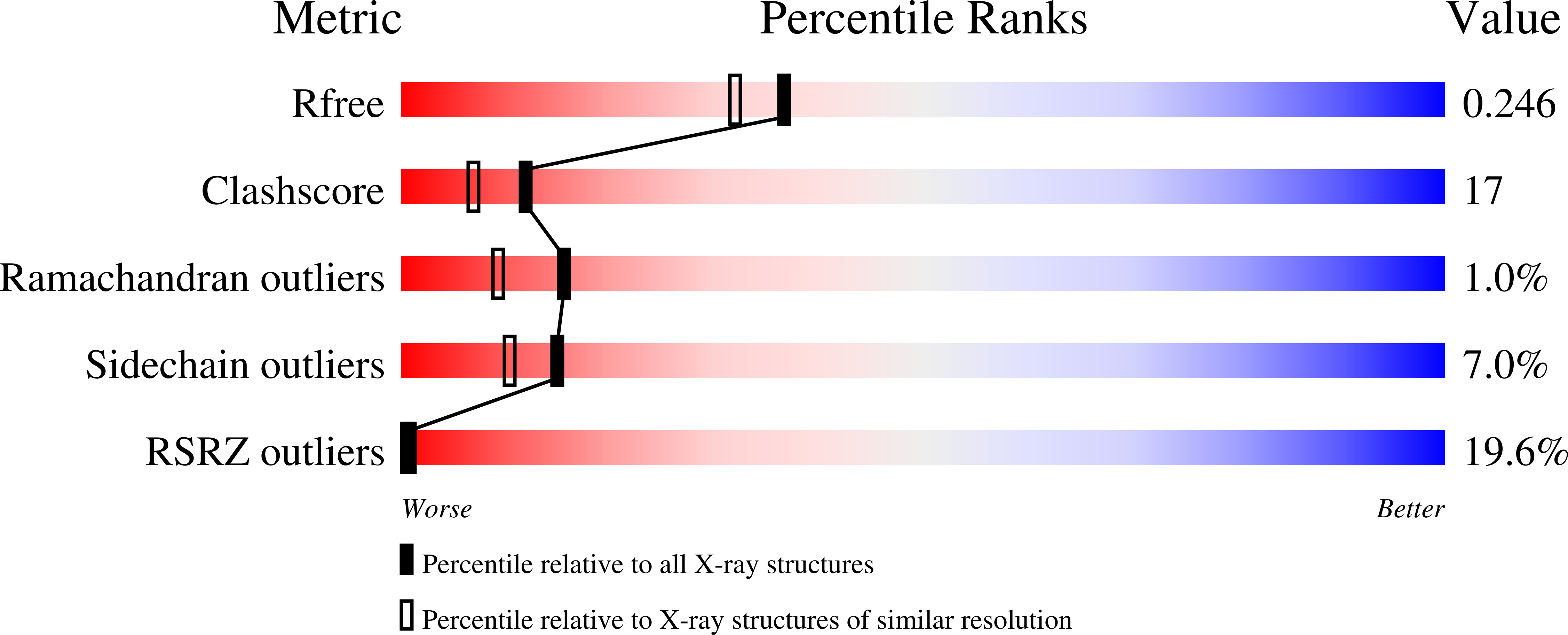
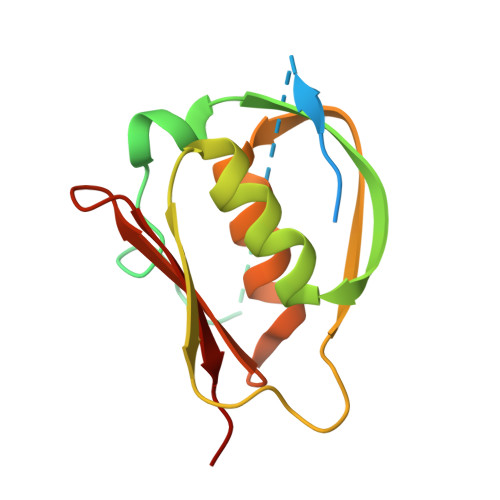
Crystal structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems
Nakano, N., Kubori, T., Kinoshita, M., Imada, K., Nagai, H.(2010) PLoS Pathog 6: e1001129-e1001129
- PubMed: 20949065
- DOI: https://doi.org/10.1371/journal.ppat.1001129
- Primary Citation of Related Structures:
3ADY - PubMed Abstract:
The Dot/Icm type IVB secretion system (T4BSS) is a pivotal determinant of Legionella pneumophila pathogenesis. L. pneumophila translocate more than 100 effector proteins into host cytoplasm using Dot/Icm T4BSS, modulating host cellular functions to establish a replicative niche within host cells. The T4BSS core complex spanning the inner and outer membranes is thought to be made up of at least five proteins: DotC, DotD, DotF, DotG and DotH. DotH is the outer membrane protein; its targeting depends on lipoproteins DotC and DotD. However, the core complex structure and assembly mechanism are still unknown. Here, we report the crystal structure of DotD at 2.0 Å resolution. The structure of DotD is distinct from that of VirB7, the outer membrane lipoprotein of the type IVA secretion system. In contrast, the C-terminal domain of DotD is remarkably similar to the N-terminal subdomain of secretins, the integral outer membrane proteins that form substrate conduits for the type II and the type III secretion systems (T2SS and T3SS). A short β-segment in the otherwise disordered N-terminal region, located on the hydrophobic cleft of the C-terminal domain, is essential for outer membrane targeting of DotH and Dot/Icm T4BSS core complex formation. These findings uncover an intriguing link between T4BSS and T2SS/T3SS.
Organizational Affiliation:
Research Institute for Microbial Diseases, Osaka University, Suita, Osaka, Japan.