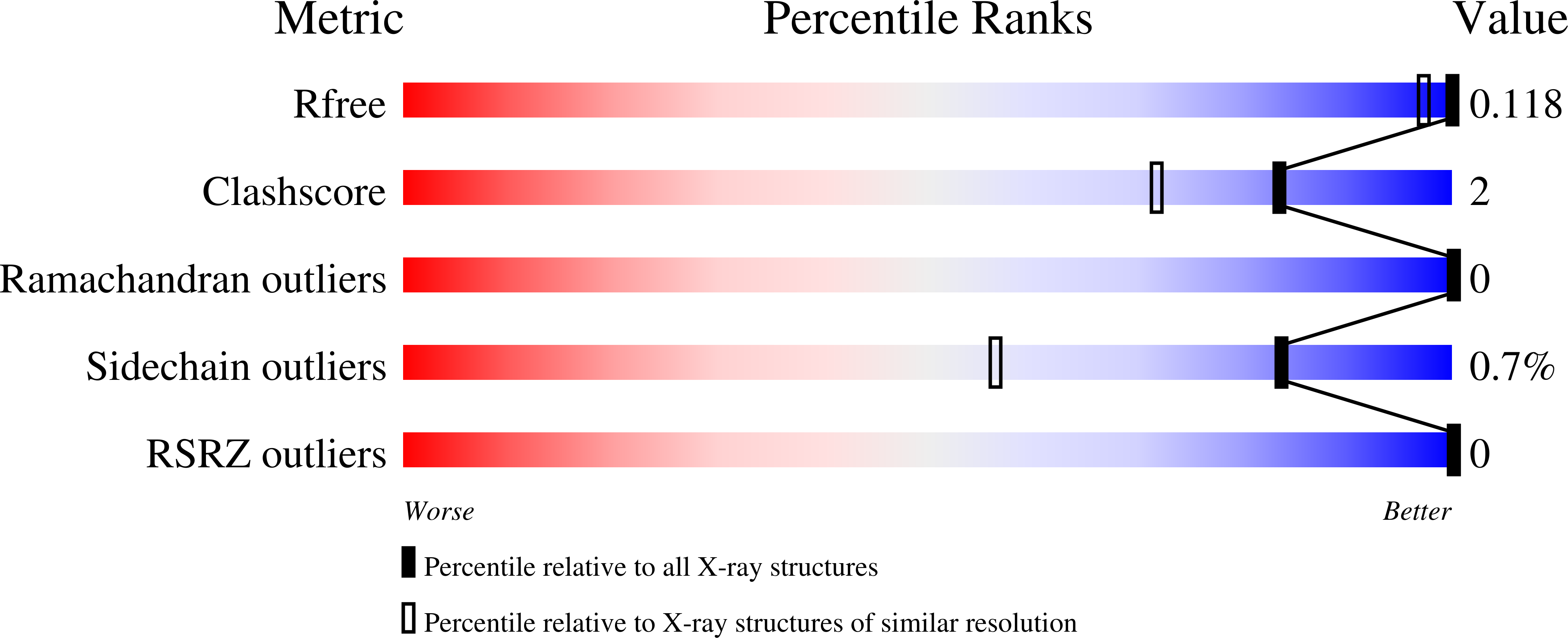
High-resolution structure of exo-arabinanase from Penicillium chrysogenum
Sogabe, Y., Kitatani, T., Yamaguchi, A., Kinoshita, T., Adachi, H., Takano, K., Inoue, T., Mori, Y., Matsumura, H., Sakamoto, T., Tada, T.(2011) Acta Crystallogr D Biol Crystallogr 67: 415-422
- PubMed: 21543843
- DOI: https://doi.org/10.1107/S0907444911006299
- Primary Citation of Related Structures:
3A71, 3A72 - PubMed Abstract:
Arabinanase Abnx from Penicillium chrysogenum 31B, which belongs to the GH93 family, releases arabinobiose from the nonreducing terminus of α-1,5-L-arabinan, which is distributed in the primary cell walls of higher plants. Crystal structures of Abnx and of its complex with arabinobiose were determined at the high resolutions of 1.14 Å to an R(work) of 10.7% (R(free) = 12.8%) and 1.04 Å to an R(work) of 10.4% (R(free) = 12.5%). Abnx has a six-bladed β-propeller fold with a typical ring-closure mode called `Velcro', in which the last four-stranded β-sheet is completed by the incorporation of a strand from the N-terminus. Catalytic residues which act as a nucleophile and an acid/base were proposed from the structures and confirmed by site-directed mutagenesis. The substrate-binding groove is enclosed at one end by two residues, Glu64 and Tyr66, which contribute to the recognition of the nonreducing chain end of the polysaccharide. A comparison with the related enzyme Arb93A which has a quite similar overall structure suggested that Abnx has different mechanisms to funnel substrates to the active site and/or to stabilize the transition state.
Organizational Affiliation:
Graduate School of Science, Osaka Prefecture University, Sakai, Osaka 599-8531, Japan.