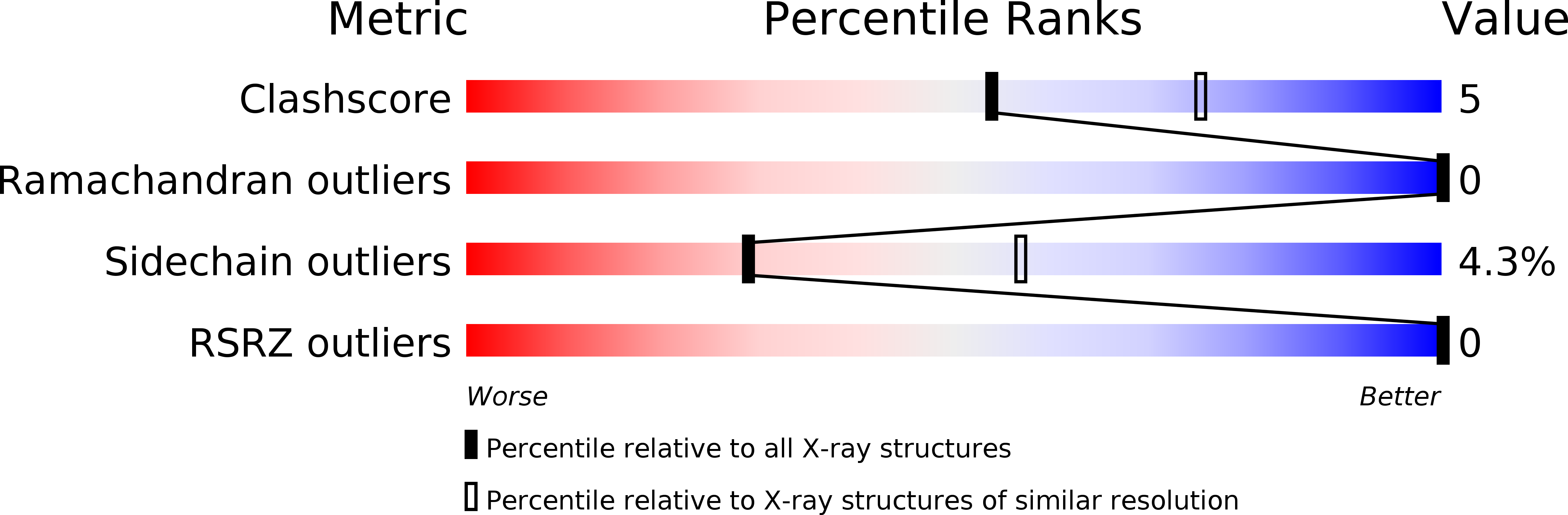
A domain flip as a result of a single amino-acid substitution.
Pokkuluri, P.R., Huang, D.B., Raffen, R., Cai, X., Johnson, G., Stevens, P.W., Stevens, F.J., Schiffer, M.(1998) Structure 6: 1067-1073
- PubMed: 9739086
- DOI: https://doi.org/10.1016/s0969-2126(98)00107-5
- Primary Citation of Related Structures:
2LVE, 3LVE, 4LVE - PubMed Abstract:
The self-assembly properties of beta domains are important features of diverse classes of proteins that include cell-adhesion molecules, surface receptors and the immunoglobulin superfamily. Immunoglobulin light-chain variable domains are well suited to the study of structural factors that determine dimerization, including how residues at the interface influence the preferred dimer arrangement. Single-site mutants of light-chain variable domain Len, designated LenQ38E and LenK30T, formed 'flipped' dimers in which one domain was rotated by about 180 degrees compared with the native protein. The dimer in the native protein is similar to that found between variable domains in Fab immunoglobulin fragments. When compared to the native dimer, more surface area is buried, and more hydrogen bonds and salt bridges are formed between the monomers in the flipped conformation. Immunoglobulin light-chain variable domains can form a minimum of two distinct quaternary structures. Single-site mutations resulting from changes of one base, such as the exchange of Gln38 to Glu or Lys30 to Thr, change the 'conventional' dimer of protein Len to a flipped arrangement. Native Len is not found in the flipped-domain dimer conformation because it would have excess positive electrostatic potential at the dimer interface that is not compensated by other forces. Excess negative or positive electrostatic potential at the dimer interface can have a determining effect on the mode of dimerization.
Organizational Affiliation:
Center for Mechanistic Biology and Biotechnology, Argonne National Laboratory, IL 60439, USA.