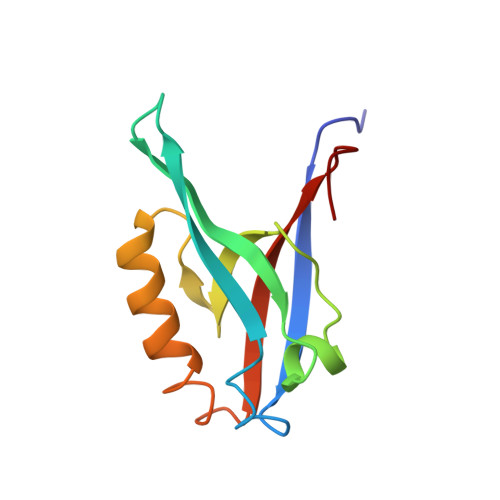
Distal interactions within the par3-VE-cadherin complex.
Tyler, R.C., Peterson, F.C., Volkman, B.F.(2010) Biochemistry 49: 951-957
- PubMed: 20047332
- DOI: https://doi.org/10.1021/bi9017335
- Primary Citation of Related Structures:
2KOH - PubMed Abstract:
par3 is a multiple-PDZ-containing scaffold protein that is central to the organization of an evolutionarily conserved cell polarity complex consisting of par3, par6, and aPKC. The ability of par3 PDZ domains to target various adhesion molecules and enzymes at the plasma membrane leads to the controlled localization of par6 and aPKC, which has firmly established its role in epithelial cell polarity. Of the numerous PDZ ligands associated with par3, interaction of its third PDZ domain with the class II ligand found within the C-terminal tail of vascular endothelial cadherin (VE-Cad) suggests a role in endothelial cell polarity as well, but the molecular details of the interaction are unknown. Previously determined structures of par3-PDZ3 bound to the class I ligand found within the C-terminal tail of the phosphoinositide phosphatase PTEN revealed two discrete binding sites: a canonical PDZ-ligand interaction site and a distal site involving charge-charge complements. Currently, it is unclear if par3-PDZ3 employs both canonical and distal binding modes in its association with VE-Cad or if these modes are unique to the PTEN interaction, suggesting a possible mechanism for ligand specificity within the polarity network. The structure of par3-PDZ3 bound to the C-terminal tail of VE-Cad presented in this work shows that both canonical and distal interactions are utilized in binding. Biophysical measurements using fluorescence polarization and two-dimensional NMR implicate the intermolecular charge pairing of aspartic acid 777 (VE-Cad) and arginine 609 (par3-PDZ3) as a crucial modulator of complex formation. Phosphorylation of VE-Cad at serine 776 increases its affinity for par3, demonstrating that post-translational modifications outside of the canonical carboxylate binding site can enhance PDZ-ligand interactions. Comparison of the VE-Cad and PTEN complexes highlights how the unique molecular architecture of par3-PDZ3 can accommodate both canonical and distal interaction modes that allow dual-class specificity for these two ligand types.
Organizational Affiliation:
Department of Biochemistry, Medical College of Wisconsin, Milwaukee, Wisconsin 53226, USA.