Structural Basis of P63Alpha Sam Domain Mutants Involved in Aec Syndrome.
Sathyamurthy, A., Freund, S.M.V., Johnson, C.M., Allen, M.D., Bycroft, M.(2011) FEBS J 278: 2680
- PubMed: 21615690
- DOI: https://doi.org/10.1111/j.1742-4658.2011.08194.x
- Primary Citation of Related Structures:
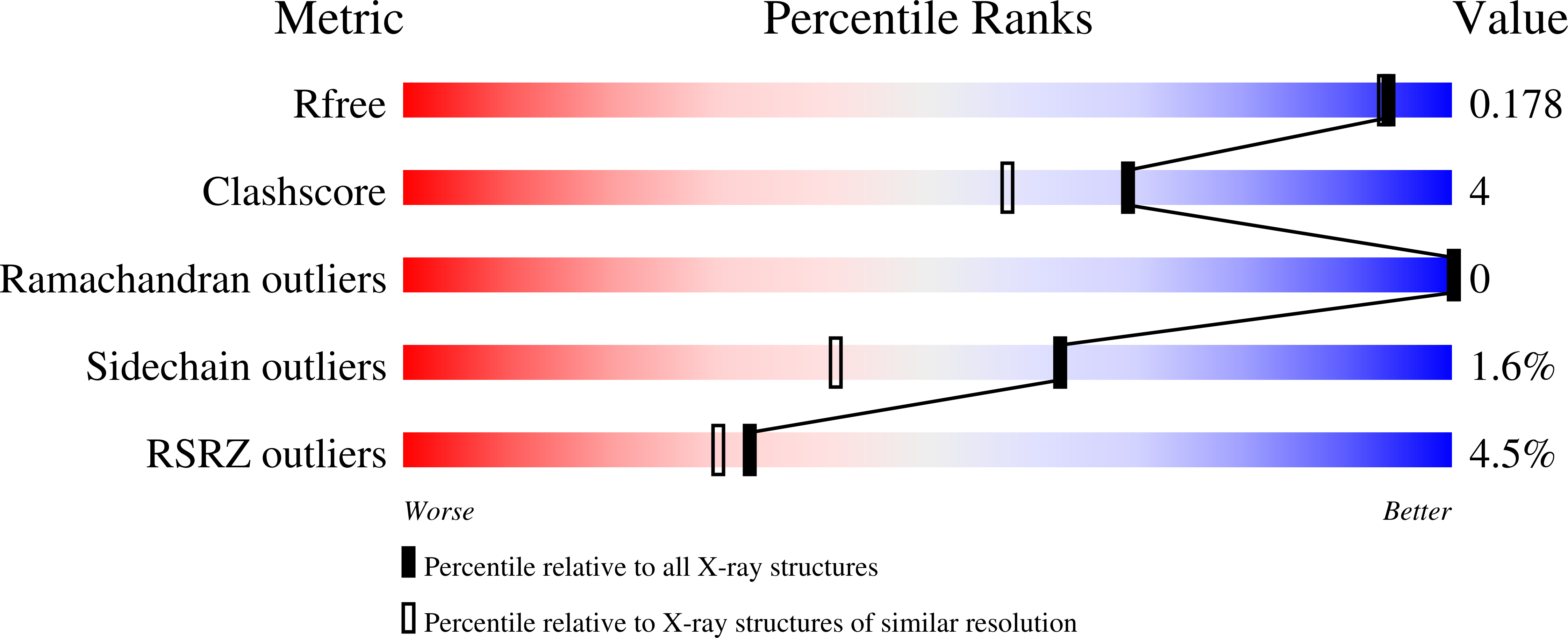
2Y9T, 2Y9U - PubMed Abstract:
p63 is a member of the p53 tumour suppressor family that includes p73. The p63 gene encodes a protein comprising an N-terminal transactivation domain, a DNA binding domain and an oligomerization domain, but varies in the organization of the C-terminus as a result of complex alternative splicing. p63α contains a C-terminal sterile α motif (SAM) domain that is thought to function as a protein-protein interaction domain. Several missense and heterozygous frame shift mutations, encoded within exon 13 and 14 of the p63 gene, have been identified in the p63α SAM domain in patients suffering from ankyloblepharon-ectodermal dysplasia-clefting syndrome. Here we report the solution and high resolution crystal structures of the p63α SAM domain and investigate the effect of several mutations (L553F/V, C562G/W, G569V, Q575L and I576T) on the stability of the domain. The possible effects of other mutations are also discussed.
Organizational Affiliation:
MRC Centre for Protein Engineering, Cambridge, UK.