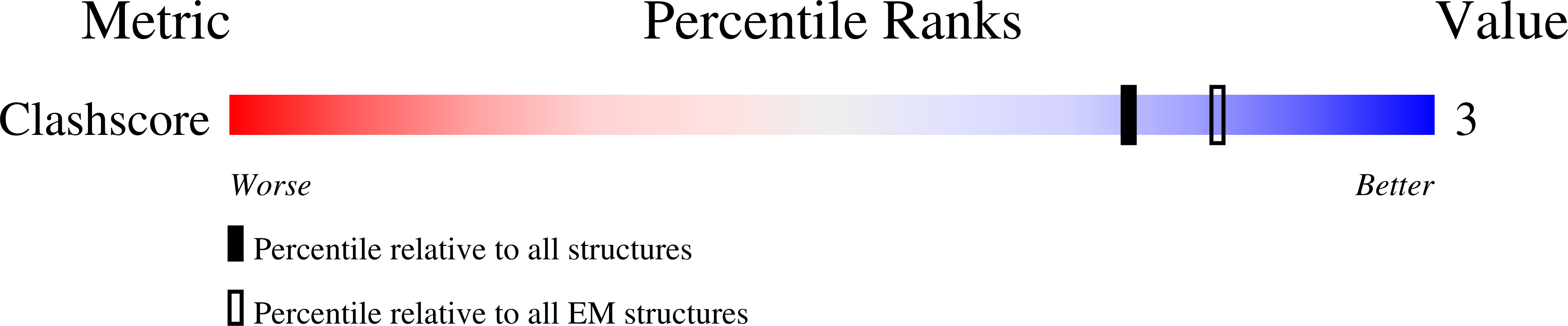Self-Directed Assembly and Clustering of the Cytoplasmic Domains of Inwardly Rectifying Kir2.1 Potassium Channels on Association with Psd-95
Fomina, S., Howard, T.D., Sleator, O.K., Golovanova, M., O'Ryan, L., Leyland, M.L., Grossmann, J.G., Collins, R.F., Prince, S.M.(2011) Biochim Biophys Acta 1808: 2374
- PubMed: 21756874
- DOI: https://doi.org/10.1016/j.bbamem.2011.06.021
- Primary Citation of Related Structures:
2XKX, 2XKY - PubMed Abstract:
The interaction of the extra-membranous domain of tetrameric inwardly rectifying Kir2.1 ion channels (Kir2.1NC(4)) with the membrane associated guanylate kinase protein PSD-95 has been studied using Transmission Electron Microscopy in negative stain. Three types of complexes were observed in electron micrographs corresponding to a 1:1 complex, a large self-enclosed tetrad complex and extended chains of linked channel domains. Using models derived from small angle X-ray scattering experiments in which high resolution structures from X-ray crystallographic and Nuclear Magnetic Resonance studies are positioned, the envelopes from single particle analysis can be resolved as a Kir2.1NC(4):PSD-95 complex and a tetrad of this unit (Kir2.1NC(4):PSD-95)(4). The tetrad complex shows the close association of the Kir2.1 cytoplasmic domains and the influence of PSD-95 mediated self-assembly on the clustering of these channels.
Organizational Affiliation:
University of Manchester, Manchester Interdisciplinary Biocentre, Manchester M1 7DN, UK.