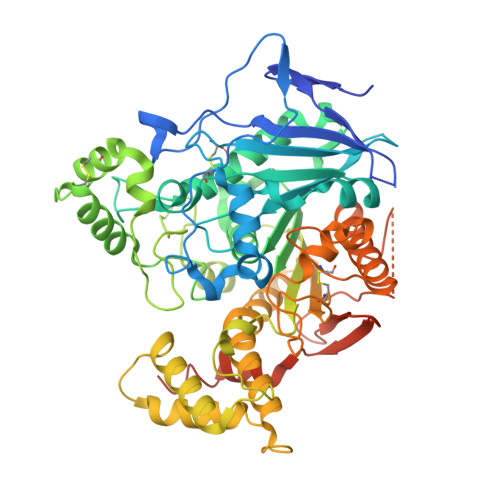
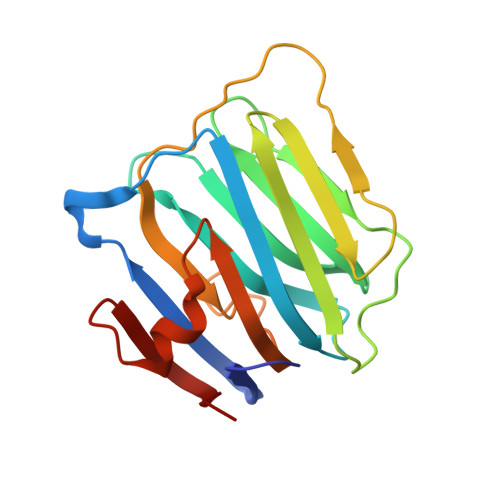
Structural Insights Into the Exquisite Selectivity of Neurexin-Neuroligin Synaptic Interactions
Leone, P., Comoletti, D., Ferracci, G., Conrod, S., Garcia, S.U., Taylor, P., Bourne, Y., Marchot, P.(2010) EMBO J 29: 2461
- PubMed: 20543817
- DOI: https://doi.org/10.1038/emboj.2010.123
- Primary Citation of Related Structures:
2XB6 - PubMed Abstract:
The extracellular domains of neuroligins and neurexins interact through Ca(2+) to form flexible trans-synaptic associations characterized by selectivity for neuroligin or neurexin subtypes. This heterophilic interaction, essential for synaptic maturation and differentiation, is regulated by gene selection, alternative mRNA splicing and post-translational modifications. A new, 2.6 A-resolution crystal structure of a soluble neurexin-1beta-neuroligin-4 (Nrx1beta-NL4) complex permits a detailed description of the Ca(2+)-coordinated interface and unveils concerted positional rearrangements of several residues of NL4, not observed in neuroligin-1, associated with Nrx1beta binding. Surface plasmon resonance analysis of the binding of structure-guided Nrx1beta mutants towards NL4 and neuroligin-1 shows that flexibility of the Nrx1beta-binding site in NL4 is reflected in a greater dissociation constant of the complex and higher sensitivity to ionic strength and pH variations. Analysis of neuroligin mutants points to critical functions for two respective residues in neuroligin-1 and neuroligin-2 in governing the affinity of the complexes. Although neuroligin-1 and neuroligin-2 have pre-determined conformations that respectively promote and prevent Nrx1beta association, unique conformational reshaping of the NL4 surface is required to permit Nrx1beta association.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, CNRS/Université d'Aix-Marseille, Campus Luminy, Marseille, France.