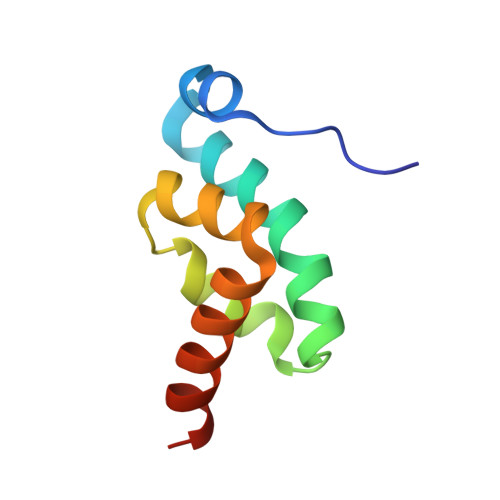
Structural Insights Into the Human Gw182-Pabc Interaction in Microrna-Mediated Deadenylation
Jinek, M., Fabian, M.R., Coyle, S.M., Sonenberg, N., Doudna, J.A.(2010) Nat Struct Mol Biol 17: 238
- PubMed: 20098421
- DOI: https://doi.org/10.1038/nsmb.1768
- Primary Citation of Related Structures:
2X04 - PubMed Abstract:
GW182-family proteins are essential for microRNA-mediated translational repression and deadenylation in animal cells. Here we show that a conserved motif in the human GW182 paralog TNRC6C interacts with the C-terminal domain of polyadenylate binding protein 1 (PABC) and present the crystal structure of the complex. Mutations at the complex interface impair mRNA deadenylation in mammalian cell extracts, suggesting that the GW182-PABC interaction contributes to microRNA-mediated gene silencing.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of California, Berkeley, California, USA.