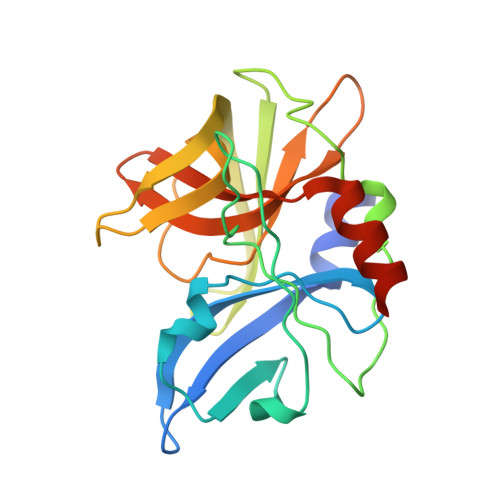
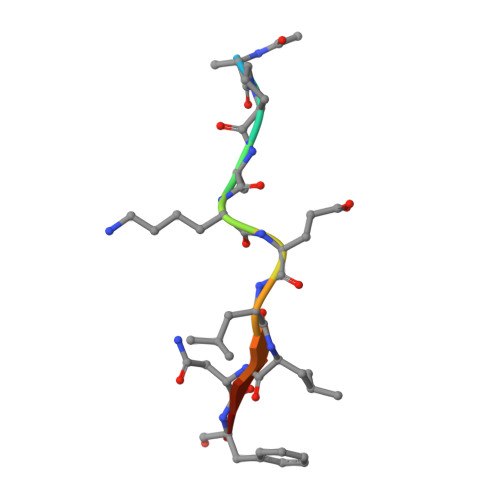
Insights Into Cleavage Specificity from the Crystal Structure of Foot-and-Mouth Disease Virus 3C Protease Complexed with a Peptide Substrate.
Zunszain, P.A., Knox, S.R., Sweeney, T.R., Yang, J., Roque-Rosell, N., Belsham, G.J., Leatherbarrow, R.J., Curry, S.(2010) J Mol Biol 395: 375
- PubMed: 19883658
- DOI: https://doi.org/10.1016/j.jmb.2009.10.048
- Primary Citation of Related Structures:
2WV4, 2WV5 - PubMed Abstract:
Picornavirus replication is critically dependent on the correct processing of a polyprotein precursor by 3C protease(s) (3C(pro)) at multiple specific sites with related but non-identical sequences. To investigate the structural basis of its cleavage specificity, we performed the first crystallographic structural analysis of non-covalent complexes of a picornavirus 3C(pro) with peptide substrates. The X-ray crystal structure of the foot-and-mouth disease virus 3C(pro), mutated to replace the catalytic Cys by Ala and bound to a peptide (APAKQ|LLNFD) corresponding to the P5-P5' region of the VP1-2A cleavage junction in the viral polyprotein, was determined up to 2.5 A resolution. Comparison with free enzyme reveals significant conformational changes in 3C(pro) on substrate binding that lead to the formation of an extended interface of contact primarily involving the P4-P2' positions of the peptide. Strikingly, the deep S1' specificity pocket needed to accommodate P1'-Leu only forms when the peptide binds. Substrate specificity was investigated using peptide cleavage assays to show the impact of amino acid substitutions within the P5-P4' region of synthetic substrates. The structure of the enzyme-peptide complex explains the marked substrate preferences for particular P4, P2 and P1 residue types, as well as the relative promiscuity at P3 and on the P' side of the scissile bond. Furthermore, crystallographic analysis of the complex with a modified VP1-2A peptide (APAKE|LLNFD) containing a Gln-to-Glu substitution reveals an identical mode of peptide binding and explains the ability of foot-and-mouth disease virus 3C(pro) to cleave sequences containing either P1-Gln or P1-Glu. Structure-based mutagenesis was used to probe interactions within the S1' specificity pocket and to provide direct evidence of the important contribution made by Asp84 of the Cys-His-Asp catalytic triad to proteolytic activity. Our results provide a new level of detail in our understanding of the structural basis of polyprotein cleavage by 3C(pro).
Organizational Affiliation:
Biophysics Section, Blackett Laboratory, Imperial College, Exhibition Road, London SW7 2AZ, UK.