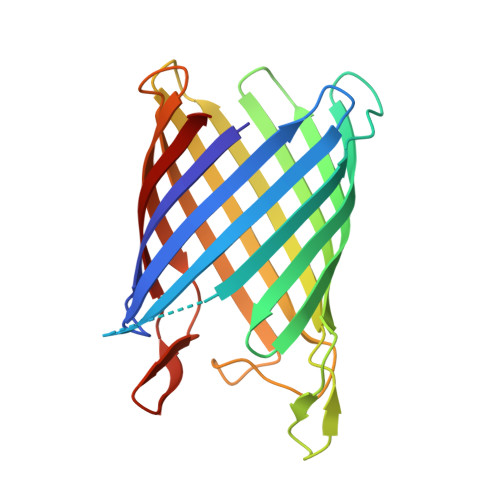
Nanc Crystal Structure, a Model for Outer Membrane Channels of the Acidic Sugar-Specific Kdgm Porin Family.
Wirth, C., Condemine, G., Boiteux, C., Berneche, S., Schirmer, T., Peneff, C.M.(2009) J Mol Biol 394: 718
- PubMed: 19796645
- DOI: https://doi.org/10.1016/j.jmb.2009.09.054
- Primary Citation of Related Structures:
2WJQ, 2WJR - PubMed Abstract:
Sialic acids are acidic sugars present mostly on vertebrate cell surfaces, which can be metabolized by bacteria and act as an inflammation signal. N-Acetylneuraminic acid, the most abundant sialic acid, can enter into Escherichia coli K12 through NanC, an N-acetylneuraminic acid-inducible outer-membrane channel. With its 215 residues, NanC belongs to the family of small monomeric KdgM-related porins. KdgM homologues are found in gammaproteobacteria, including major plant and human pathogens, and together they define a large family of putative acidic sugar/oligosaccharide transporters, which are as yet poorly characterized. Here, we present the first high-resolution structure of a KdgM family member. NanC folds into a 28-A-high, 12-stranded beta-barrel, resembling the beta-domain of autotransporter NalP and defining an open pore with an average radius of 3.3 A. The channel is lined by two strings of basic residues facing each other across the pore, a feature that appears largely conserved within the KdgM family and is likely to facilitate the diffusion of acidic oligosaccharides.
Organizational Affiliation:
Department of Structural Biology, Biozentrum, University of Basel, Klingelbergstrasse 70, CH-4056 Basel, Switzerland.