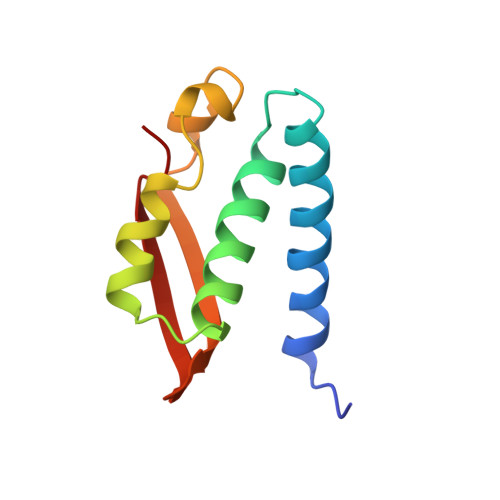
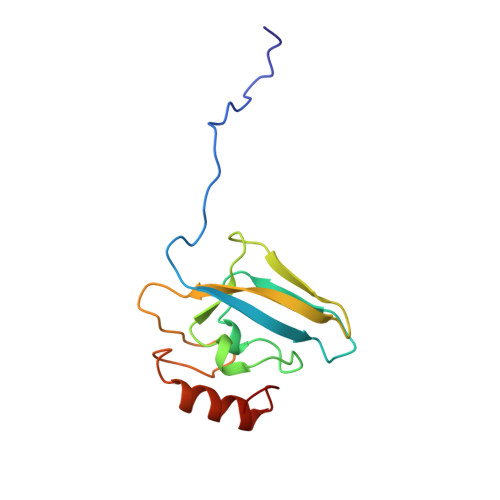
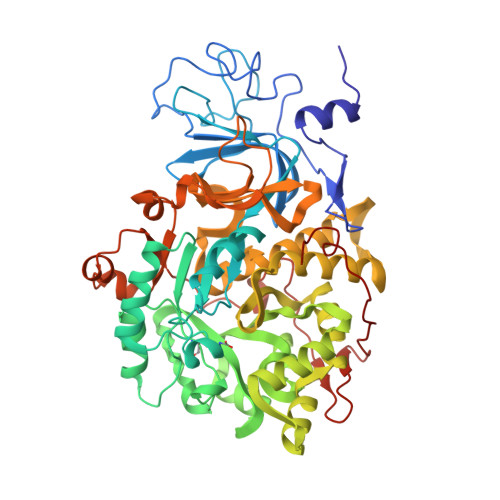
A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels.
Benini, S., Rypniewski, W.R., Wilson, K.S., Miletti, S., Ciurli, S., Mangani, S.(1999) Structure 7: 205-216
- PubMed: 10368287
- DOI: https://doi.org/10.1016/S0969-2126(99)80026-4
- Primary Citation of Related Structures:
2UBP, 3UBP - PubMed Abstract:
Urease catalyzes the hydrolysis of urea, the final step of organic nitrogen mineralization, using a bimetallic nickel centre. The role of the active site metal ions and amino acid residues has not been elucidated to date. Many pathologies are associated with the activity of ureolytic bacteria, and the efficiency of soil nitrogen fertilization with urea is severely decreased by urease activity. Therefore, the development of urease inhibitors would lead to a reduction of environmental pollution, to enhanced efficiency of nitrogen uptake by plants, and to improved therapeutic strategies for treatment of infections due to ureolytic bacteria. Structure-based design of urease inhibitors would require knowledge of the enzyme mechanism at the molecular level.
Organizational Affiliation:
European Molecular Biology Laboratory, DESY, Notkestrasse 85 D-22603, Hamburg, Germany.