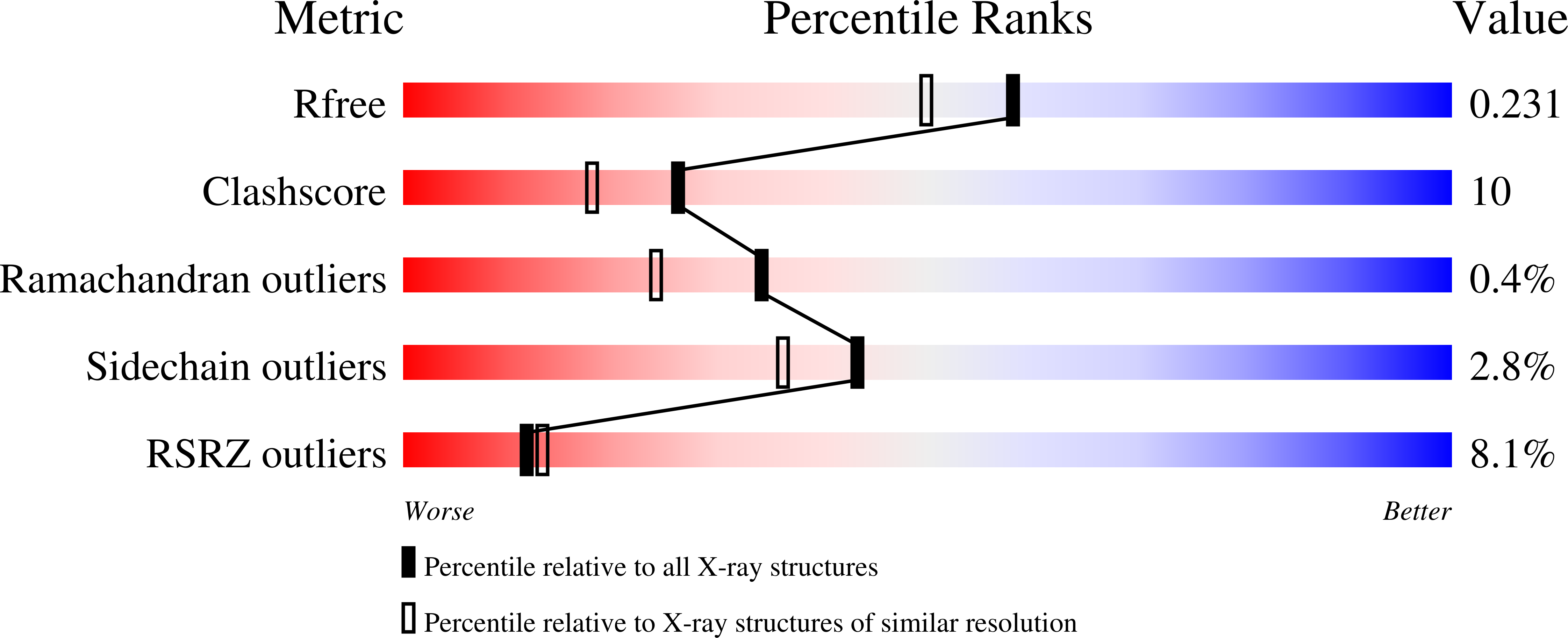
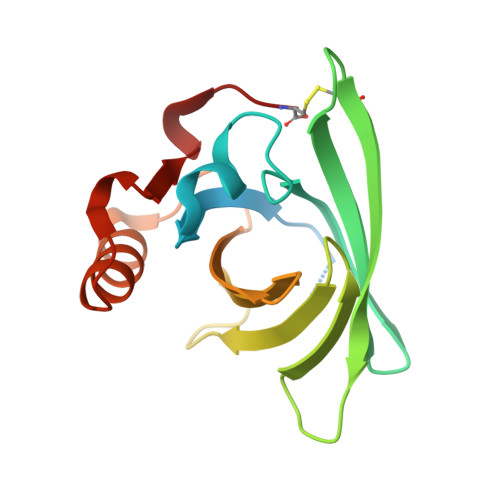
Three-dimensional structure and ligand binding properties of trichosurin, a metatherian lipocalin from the milk whey of the common brushtail possum Trichosurus vulpecula
Watson, R.P., Demmer, J., Baker, E.N., Arcus, V.L.(2007) Biochem J 408: 29-38
- PubMed: 17685895
- DOI: https://doi.org/10.1042/BJ20070567
- Primary Citation of Related Structures:
2R73, 2R74, 2RA6 - PubMed Abstract:
Lipocalins are extracellular proteins (17-25 kDa) that bind and transport small lipophilic molecules. The three-dimensional structure of the first lipocalin from a metatherian has been determined at different values of pH both with and without bound ligands. Trichosurin, a protein from the milk whey of the common brushtail possum, Trichosurus vulpecula, has been recombinantly expressed in Escherichia coli, refolded from inclusion bodies, purified and crystallized at two different pH values. The three-dimensional structure of trichosurin was solved by X-ray crystallography in two different crystal forms to 1.9 A (1 A=0.1 nm) and 2.6 A resolution, from crystals grown at low and high pH values respectively. Trichosurin has the typical lipocalin fold, an eight-stranded anti-parallel beta-barrel but dimerizes in an orientation that has not been seen previously. The putative binding pocket in the centre of the beta-barrel is well-defined in both high and low pH structures and is occupied by water molecules along with isopropanol molecules from the crystallization medium. Trichosurin was also co-crystallized with a number of small molecule ligands and structures were determined with 2-naphthol and 4-ethylphenol bound in the centre of the beta-barrel. The binding of phenolic compounds by trichosurin provides clues to the function of this important marsupial milk protein, which is highly conserved across metatherians.
Organizational Affiliation:
Laboratory of Structural Biology, School of Biological Sciences, University of Auckland, Private Bag 92-019, Auckland, New Zealand.