Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition.
Janssen, B.J., Halff, E.F., Lambris, J.D., Gros, P.(2007) J Biol Chem 282: 29241-29247
- PubMed: 17684013
- DOI: https://doi.org/10.1074/jbc.M704587200
- Primary Citation of Related Structures:
2QKI - PubMed Abstract:
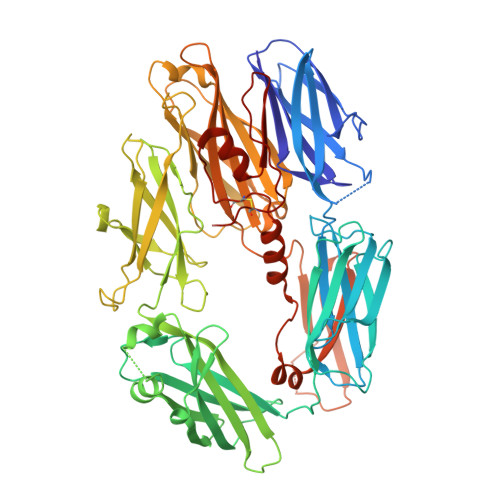
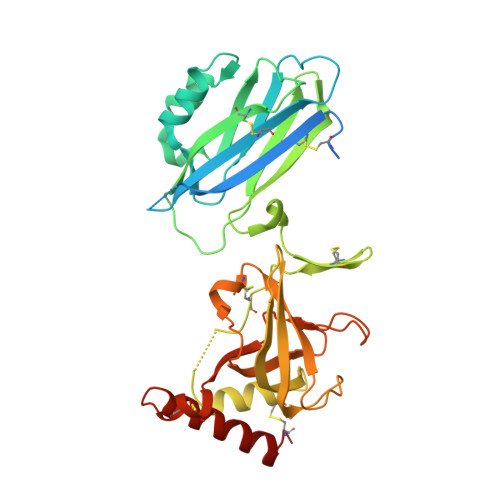
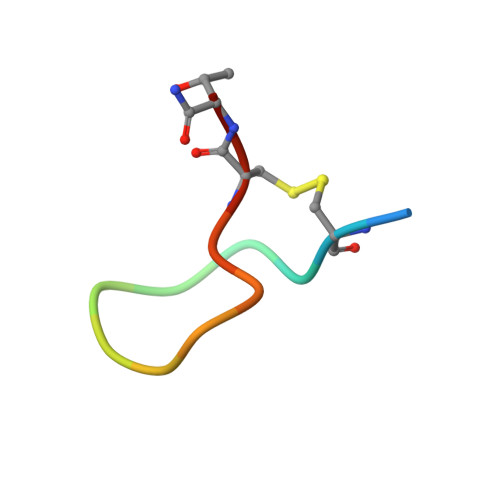
Undesired complement activation is a major cause of tissue injury in various pathological conditions and contributes to several immune complex diseases. Compstatin, a 13-residue peptide, is an effective inhibitor of the activation of complement component C3 and thus blocks a central and crucial step in the complement cascade. The precise binding site on C3, the structure in the bound form, and the exact mode of action of compstatin are unknown. Here we present the crystal structure of compstatin in complex with C3c, a major proteolytic fragment of C3. The structure reveals that the compstatin-binding site is formed by the macroglobulin (MG) domains 4 and 5. This binding site is part of the structurally stable MG-ring formed by domains MG 1-6 and is far away from any other known binding site on C3. Compstatin does not alter the conformation of C3c, whereas compstatin itself undergoes a large conformational change upon binding. We propose a model in which compstatin sterically hinders the access of the substrate C3 to the convertase complexes, thus blocking complement activation and amplification. These insights are instrumental for further development of compstatin as a potential therapeutic.
Organizational Affiliation:
Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Sciences, Utrecht University, 3584 CH Utrecht, The Netherlands.