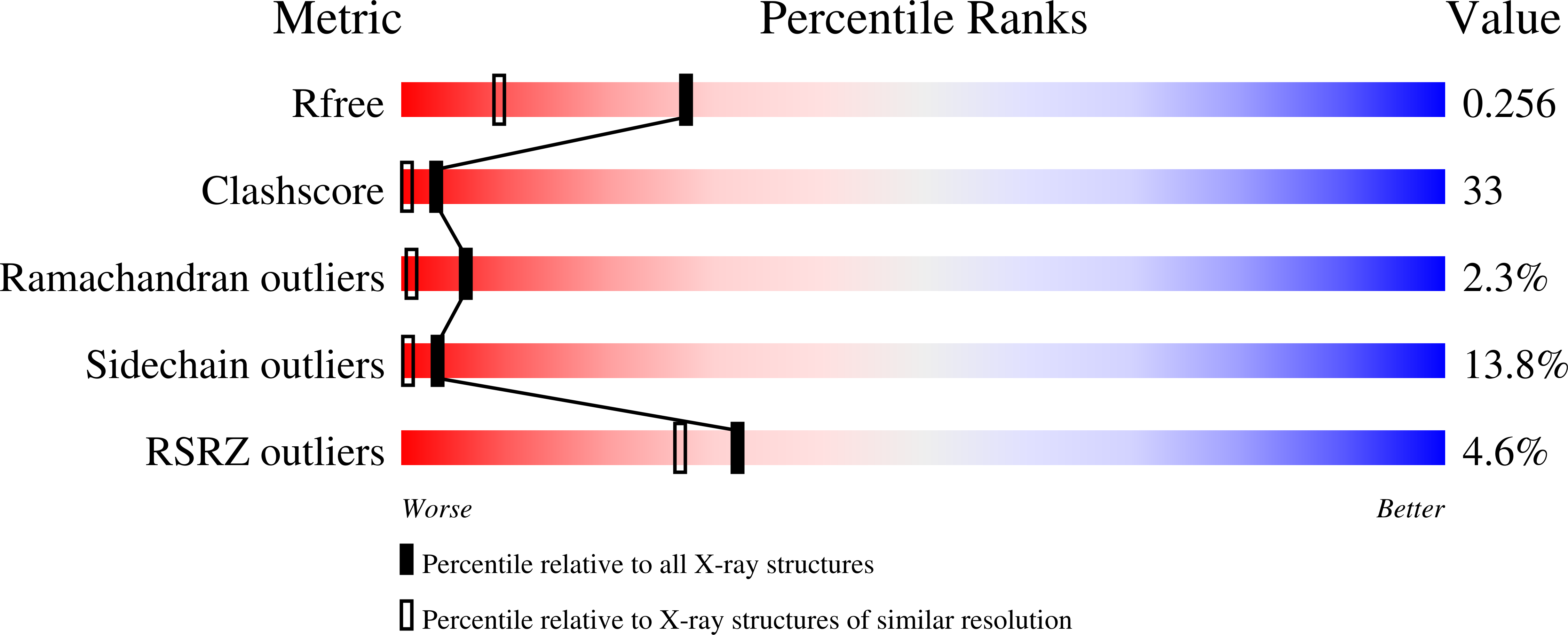
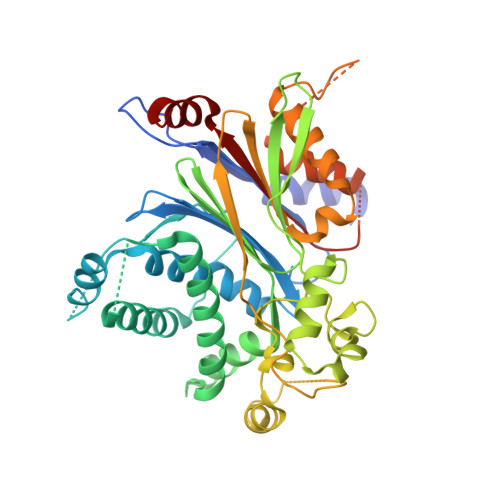
Crystal structure of pyruvate dehydrogenase phosphatase 1 and its functional implications.
Vassylyev, D.G., Symersky, J.(2007) J Mol Biol 370: 417-426
- PubMed: 17532339
- DOI: https://doi.org/10.1016/j.jmb.2007.05.002
- Primary Citation of Related Structures:
2PNQ - PubMed Abstract:
Pyruvate dehydrogenase phosphatase 1 (PDP1) catalyzes dephosphorylation of pyruvate dehydrogenase (E1) in the mammalian pyruvate dehydrogenase complex (PDC), whose activity is regulated by the phosphorylation-dephosphorylation cycle by the corresponding protein kinases (PDHKs) and phosphatases. The activity of PDP1 is greatly enhanced through Ca2+ -dependent binding of the catalytic subunit (PDP1c) to the L2 (inner lipoyl) domain of dihydrolipoyl acetyltransferase (E2), which is also integrated in PDC. Here, we report the crystal structure of the rat PDP1c at 1.8 A resolution. The structure reveals that PDP1 belongs to the PPM family of protein serine/threonine phosphatases, which, in spite of a low level of sequence identity, share the structural core consisting of the central beta-sandwich flanked on both sides by loops and alpha-helices. Consistent with the previous studies, two well-fixed magnesium ions are coordinated by five active site residues and five water molecules in the PDP1c catalytic center. Structural analysis indicates that, while the central portion of the PDP1c molecule is highly conserved among the members of the PPM protein family, a number of structural insertions and deletions located at the periphery of PDP1c likely define its functional specificity towards the PDC. One notable feature of PDP1c is a long insertion (residues 98-151) forming a unique hydrophobic pocket on the surface that likely accommodates the lipoyl moiety of the E2 domain in a fashion similar to that of PDHKs. The cavity, however, appears more open than in PDHK, suggesting that its closure may be required to achieve tight, specific binding of the lipoic acid. We propose a mechanism in which the closure of the lipoic acid binding site is triggered by the formation of the intermolecular (PDP1c/L2) Ca2+ binding site in a manner reminiscent of the Ca2+ -induced closure of the regulatory domain of troponin C.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, School of Medicine and Dentistry, Kaul Genetics Building, Birmingham, Al 35294, USA. dmitry@uab.edu