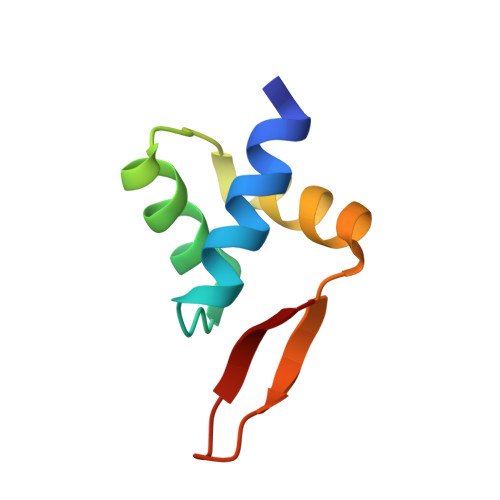
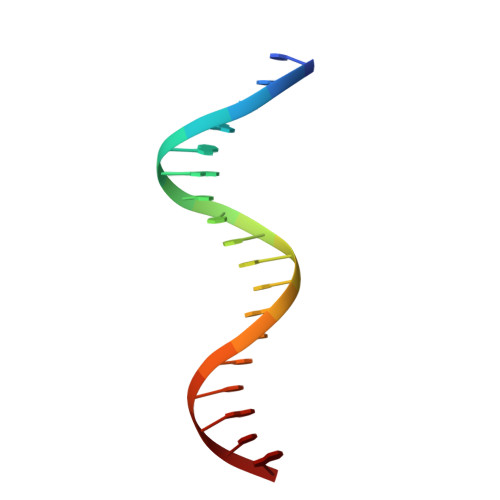
Structure and function of the arginine repressor-operator complex from Bacillus subtilis.
Garnett, J.A., Marincs, F., Baumberg, S., Stockley, P.G., Phillips, S.E.(2008) J Mol Biol 379: 284-298
- PubMed: 18455186
- DOI: https://doi.org/10.1016/j.jmb.2008.03.007
- Primary Citation of Related Structures:
2P5L - PubMed Abstract:
In many bacteria, the concentration of L-arginine is controlled by a transcriptional regulator, the arginine repressor. In Bacillus subtilis this transcription factor is called AhrC and has roles in both the repression and activation of the genes involved in arginine metabolism. It interacts with 18 bp ARG boxes in the promoters of arginine biosynthetic and catabolic operons. AhrC is a hexamer and each subunit has two domains. The C-terminal domains form the core, mediating inter-subunit interactions and L-arginine binding, while the N-terminal domains contain a winged helix-turn-helix DNA-binding motif and are arranged around the periphery. Upon binding of the co-repressor L-arginine there is a approximately 15 degrees relative rotation between core C-terminal trimers. Here, we report the X-ray crystal structure of a dimer of the N-terminal domains of AhrC (NAhrC) in complex with an 18 bp DNA ARG box operator, refined to 2.85 A resolution. Comparison of the N-terminal domains within this complex with those of the free domain reveals that the flexible beta-wings of the DNA-binding motif in the free domain form a stable dimer interface in the protein-DNA complex, favouring correct orientation of the recognition helices. These are then positioned to insert into adjacent turns of the major groove of the ARG box, whilst the wings contact the minor groove. There are extensive contacts between the protein and the DNA phosphodiester backbone, as well as a number of direct hydrogen bonds between conserved amino acid side chains and bases. Combining this structure with other crystal structures of other AhrC components, we have constructed a model of the repression complex of AhrC at the B. subtilis biosynthetic argC operator and, along with transcriptome data, analysed the origins of sequence specificity and arginine activation.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds LS2 9JT, UK.