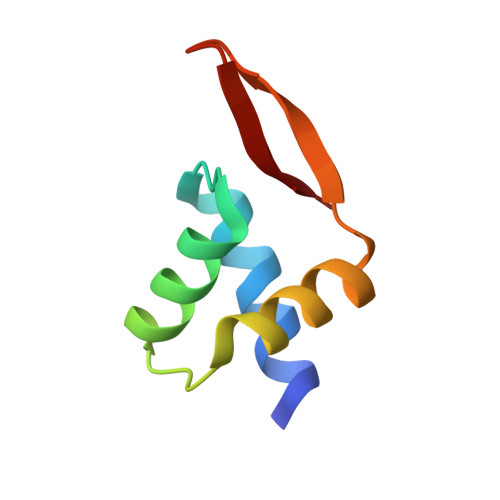
A high-resolution structure of the DNA-binding domain of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Garnett, J.A., Baumberg, S., Stockley, P.G., Phillips, S.E.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 914-917
- PubMed: 18007039
- DOI: https://doi.org/10.1107/S1744309107048166
- Primary Citation of Related Structures:
2P5K - PubMed Abstract:
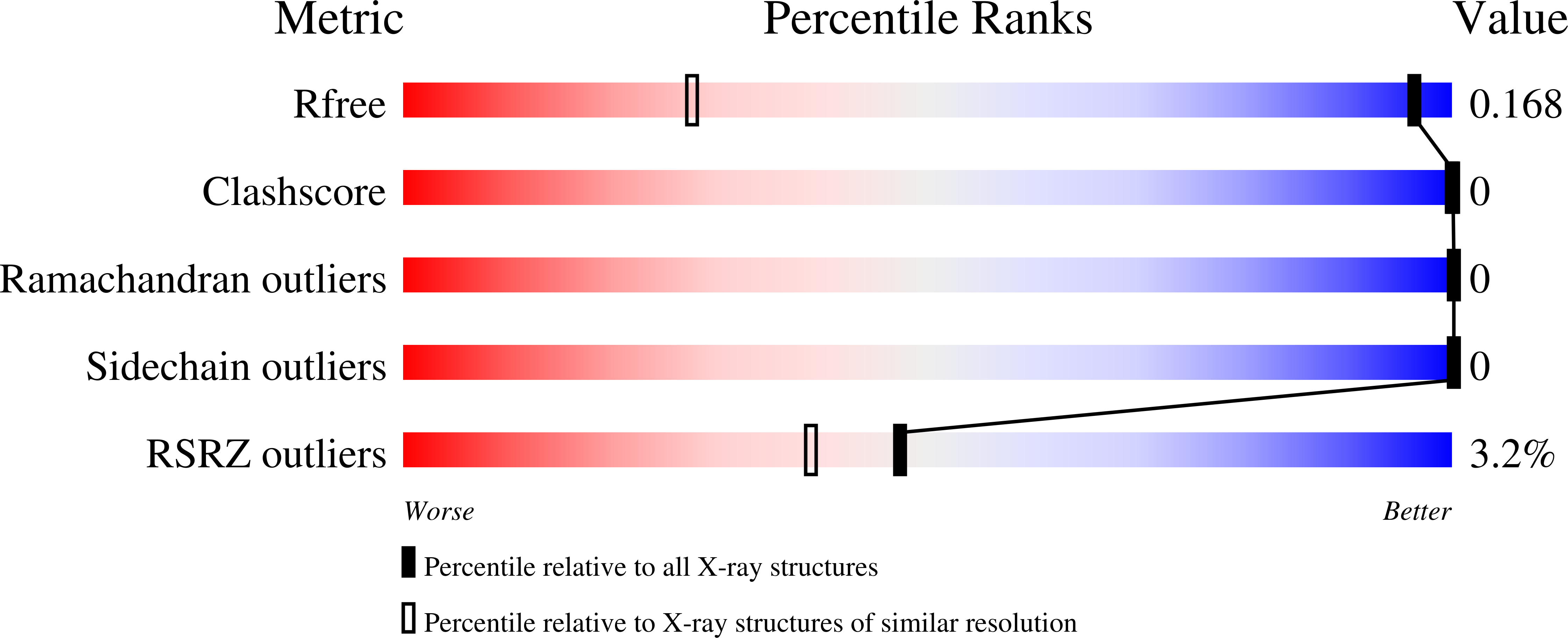
In Bacillus subtilis the concentration of L-arginine is controlled by the transcriptional regulator AhrC, which interacts with 18 bp DNA operator sites called ARG boxes in the promoters of arginine biosynthetic and catabolic operons. AhrC is a 100 kDa homohexamer, with each subunit having two domains. The C-terminal domains form the core, mediating intersubunit interactions and binding of the co-repressor L-arginine, whilst the N-terminal domains contain a winged helix-turn-helix DNA-binding motif and are arranged around the periphery. The N-terminal domain of AhrC has been expressed, purified and characterized and it has been shown that the fragment still binds DNA operators as a recombinant monomer. The DNA-binding domain has also been crystallized and the crystal structure refined to 1.0 A resolution is presented.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds LS2 9JT, England.