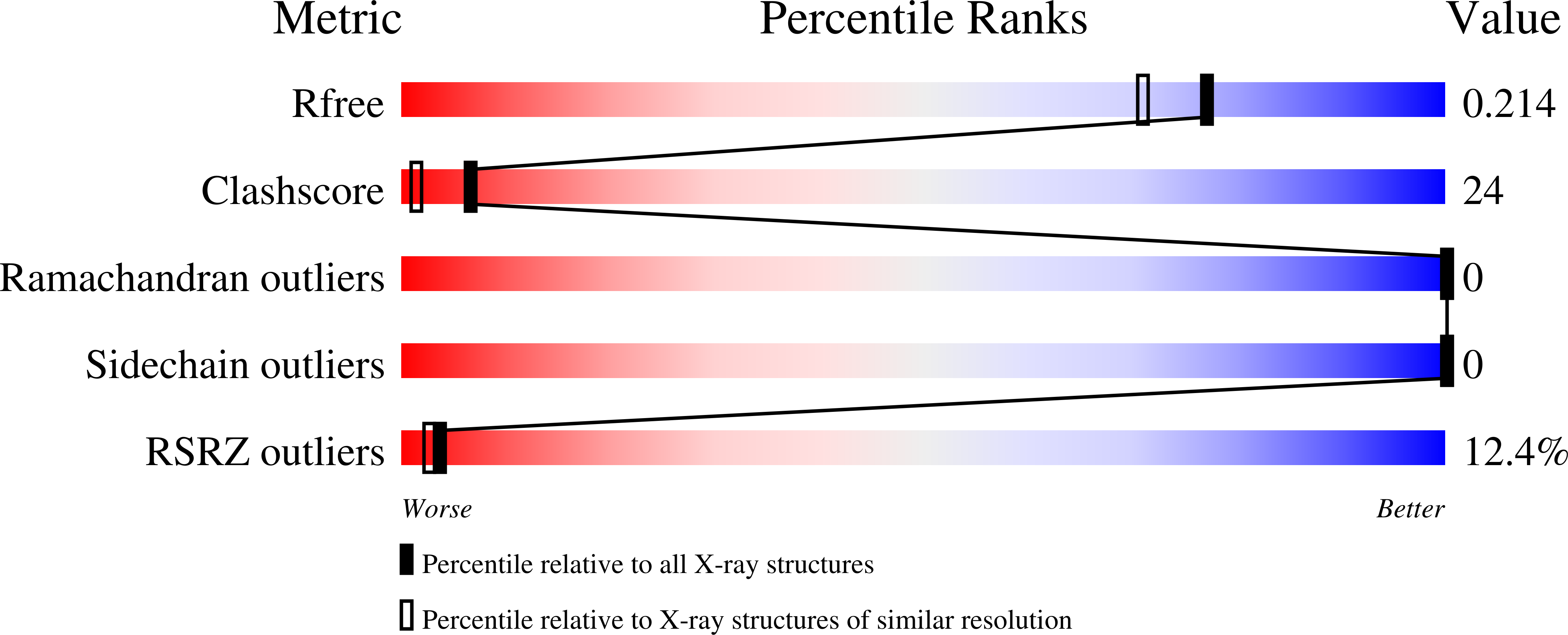
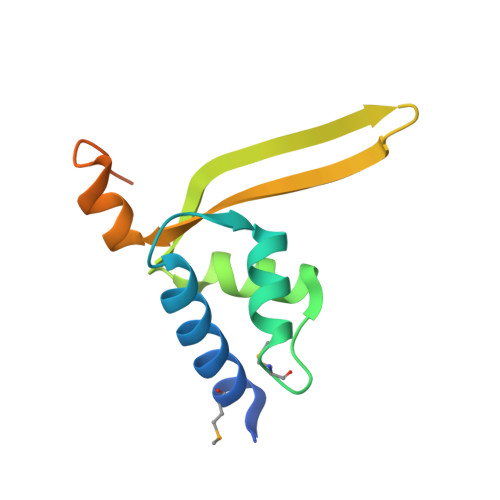
Crystal structure of hypothetical protein (JCVI_PEP_1096688149193) from an environmental metagenome (unidentified marine microbe), Sorcerer II Global Ocean Sampling experiment at 1.79 A resolution
Joint Center for Structural Genomics (JCSG)To be published.