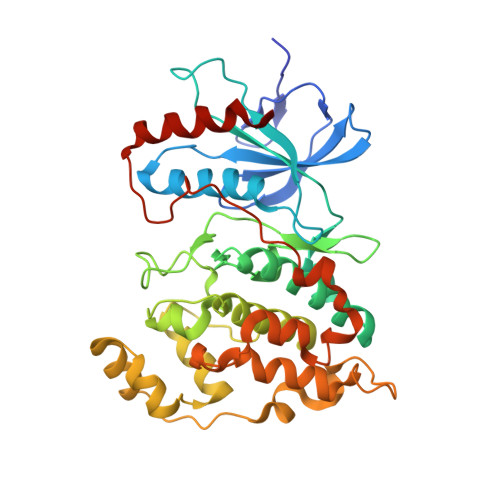
Discovery of a new class of 4-anilinopyrimidines as potent c-Jun N-terminal kinase inhibitors: Synthesis and SAR studies.
Liu, M., Wang, S., Clampit, J.E., Gum, R.J., Haasch, D.L., Rondinone, C.M., Trevillyan, J.M., Abad-Zapatero, C., Fry, E.H., Sham, H.L., Liu, G.(2007) Bioorg Med Chem Lett 17: 668-672
- PubMed: 17107797
- DOI: https://doi.org/10.1016/j.bmcl.2006.10.093
- Primary Citation of Related Structures:
2NO3 - PubMed Abstract:
A new series of 4-anilinopyrimidines has been synthesized and evaluated as JNK1 inhibitors. SAR studies led to the discovery of potent JNK1 inhibitors with good enzymatic activity as well as cellular potency represented by compound 2b. Kinase selectivity profile and the crystal structure of 2b are also described.
Organizational Affiliation:
Metabolic Disease Research, Global Pharmaceutical Research and Development, Abbott Laboratories, Abbott Park, IL 60064-6098, USA. mei.liu@abbott.com