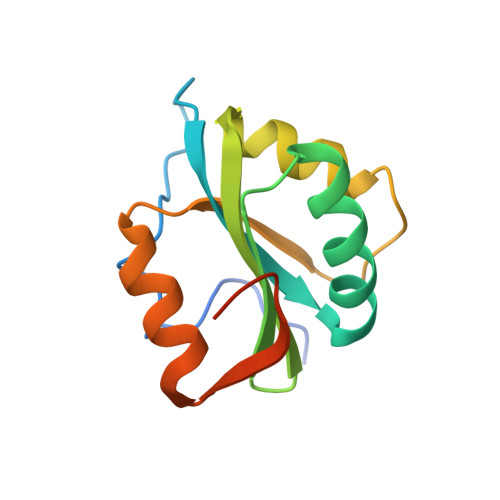
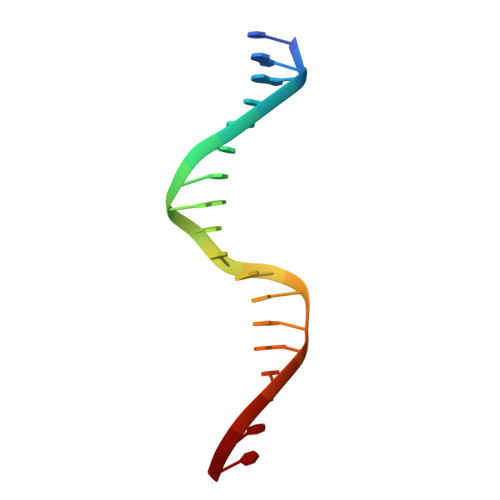
Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA
Bochkareva, E., Martynowski, D., Seitova, A., Bochkarev, A.(2006) EMBO J 25: 5961-5969
- PubMed: 17139255
- DOI: https://doi.org/10.1038/sj.emboj.7601452
- Primary Citation of Related Structures:
2IPR, 2ITJ, 2ITL, 2NL8 - PubMed Abstract:
The large T antigen (T-ag) protein binds to and activates DNA replication from the origin of DNA replication (ori) in simian virus 40 (SV40). Here, we determined the crystal structures of the T-ag origin-binding domain (OBD) in apo form, and bound to either a 17 bp palindrome (sites 1 and 3) or a 23 bp ori DNA palindrome comprising all four GAGGC binding sites for OBD. The T-ag OBDs were shown to interact with the DNA through a loop comprising Ser147-Thr155 (A1 loop), a combination of a DNA-binding helix and loop (His203-Asn210), and Asn227. The A1 loop traveled back-and-forth along the major groove and accounted for most of the sequence-determining contacts with the DNA. Unexpectedly, in both T-ag-DNA structures, the T-ag OBDs bound DNA independently and did not make direct protein-protein contacts. The T-ag OBD was also captured bound to a non-consensus site ATGGC even in the presence of its canonical site GAGGC. Our observations taken together with the known biochemical and structural features of the T-ag-origin interaction suggest a model for origin unwinding.
Organizational Affiliation:
Banting and Best Department of Medical Research & Department of Medical Genetics and Microbiology, University of Toronto, 100 College St., Toronto, Ontario, Canada.