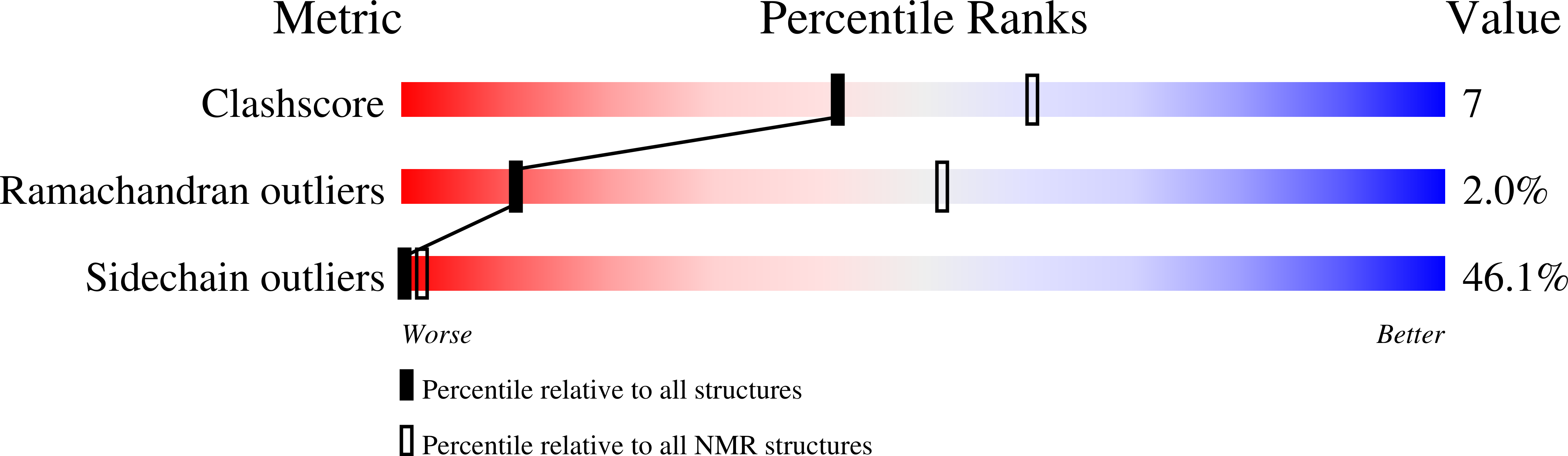Self-assembly of MPG1, a hydrophobin protein from the rice blast fungus that forms functional amyloid coatings, occurs by a surface-driven mechanism.
Pham, C.L., Rey, A., Lo, V., Soules, M., Ren, Q., Meisl, G., Knowles, T.P., Kwan, A.H., Sunde, M.(2016) Sci Rep 6: 25288-25288
- PubMed: 27142249
- DOI: https://doi.org/10.1038/srep25288
- Primary Citation of Related Structures:
2N4O - PubMed Abstract:
Rice blast is a devastating disease of rice caused by the fungus Magnaporthe oryzae and can result in loss of a third of the annual global rice harvest. Two hydrophobin proteins, MPG1 and MHP1, are highly expressed during rice blast infections. These hydrophobins have been suggested to facilitate fungal spore adhesion and to direct the action of the enzyme cutinase 2, resulting in penetration of the plant host. Therefore a mechanistic understanding of the self-assembly properties of these hydrophobins and their interaction with cutinase 2 is crucial for the development of novel antifungals. Here we report details of a study of the structure, assembly and interactions of these proteins. We demonstrate that, in vitro, MPG1 assembles spontaneously into amyloid structures while MHP1 forms a non-fibrillar film. The assembly of MPG1 only occurs at a hydrophobic:hydrophilic interface and can be modulated by MHP1 and other factors. We further show that MPG1 assemblies can much more effectively retain cutinase 2 activity on a surface after co-incubation and extensive washing compared with other protein coatings. The assembly and interactions of MPG1 and MHP1 at hydrophobic surfaces thereby provide the basis for a possible mechanism by which the fungus can develop appropriately at the infection interface.
Organizational Affiliation:
Discipline of Pharmacology, School of Medical Sciences, University of Sydney, NSW 2006, Australia.