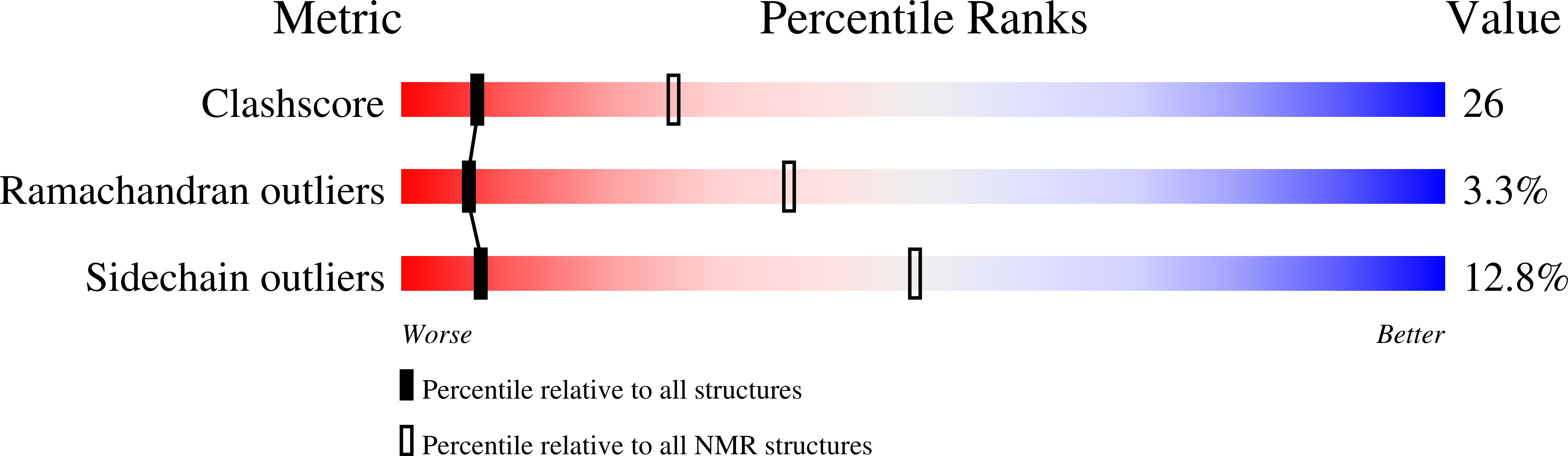Solution structure of the major factor VIII binding region on von Willebrand factor.
Shiltagh, N., Kirkpatrick, J., Cabrita, L.D., McKinnon, T.A., Thalassinos, K., Tuddenham, E.G., Hansen, D.F.(2014) Blood 123: 4143-4151
- PubMed: 24700780
- DOI: https://doi.org/10.1182/blood-2013-07-517086
- Primary Citation of Related Structures:
2MHP, 2MHQ - PubMed Abstract:
Although much of the function of von Willebrand factor (VWF) has been revealed, detailed insight into the molecular structure that enables VWF to orchestrate hemostatic processes, in particular factor VIII (FVIII) binding and stabilization in plasma, is lacking. Here, we present the high-resolution solution structure and structural dynamics of the D' region of VWF, which constitutes the major FVIII binding site. D' consists of 2 domains, trypsin-inhibitor-like (TIL') and E', of which the TIL' domain lacks extensive secondary structure, is strikingly dynamic and harbors a cluster of pathological mutations leading to decreased FVIII binding affinity (type 2N von Willebrand disease [VWD]). This indicates that the backbone malleability of TIL' is important for its biological activity. The principal FVIII binding site is localized to a flexible, positively charged region on TIL', which is supported by the rigid scaffold of the TIL' and E' domain β sheets. Furthermore, surface-charge mapping of the TIL'E' structure reveals a potential mechanism for the electrostatically guided, high-affinity VWF⋅FVIII interaction. Our findings provide novel insights into VWF⋅FVIII complex formation, leading to a greater understanding of the molecular basis of the bleeding diathesis type 2N VWD.
Organizational Affiliation:
Division of Biosciences, Institute of Structural and Molecular Biology, University College London, London, United Kingdom;