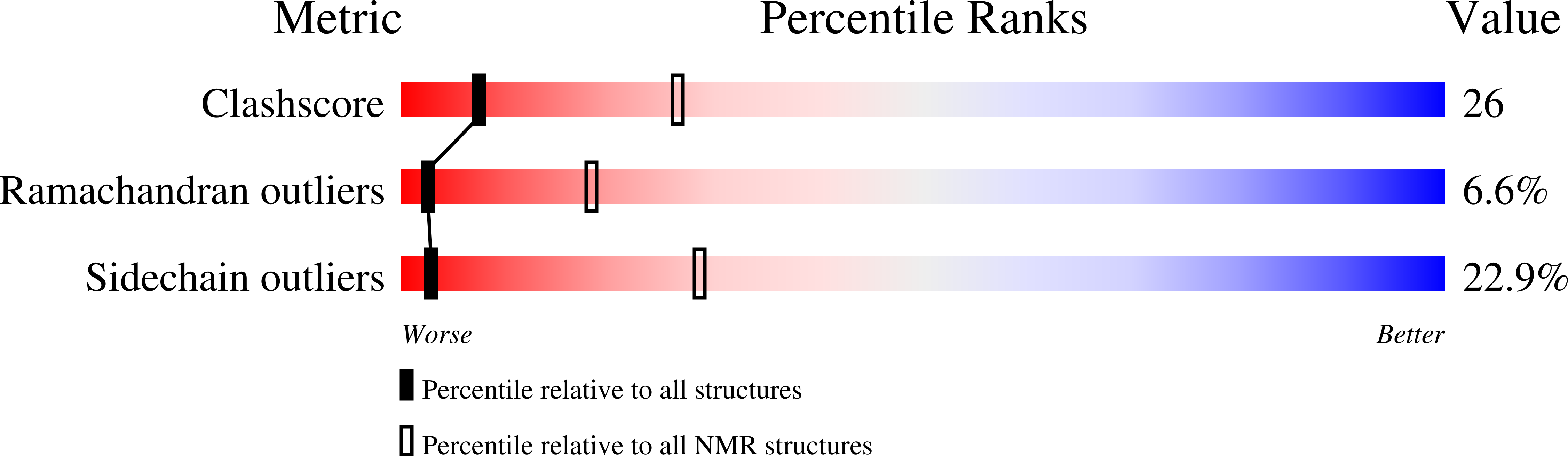A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix.
Leon, E., Navarro-Aviles, G., Santiveri, C.M., Flores-Flores, C., Rico, M., Gonzalez, C., Murillo, F.J., Elias-Arnanz, M., Jimenez, M.A., Padmanabhan, S.(2010) Nucleic Acids Res 38: 5226-5241
- PubMed: 20410074
- DOI: https://doi.org/10.1093/nar/gkq277
- Primary Citation of Related Structures:
2KSS - PubMed Abstract:
Direct targeting of critical DNA-binding elements of a repressor by its cognate antirepressor is an effective means to sequester the repressor and remove a transcription initiation block. Structural descriptions for this, though often proposed for bacterial and phage repressor-antirepressor systems, are unavailable. Here, we describe the structural and functional basis of how the Myxococcus xanthus CarS antirepressor recognizes and neutralizes its cognate repressors to turn on a photo-inducible promoter. CarA and CarH repress the carB operon in the dark. CarS, produced in the light, physically interacts with the MerR-type winged-helix DNA-binding domain of these repressors leading to activation of carB. The NMR structure of CarS1, a functional CarS variant, reveals a five-stranded, antiparallel beta-sheet fold resembling SH3 domains, protein-protein interaction modules prevalent in eukaryotes but rare in prokaryotes. NMR studies and analysis of site-directed mutants in vivo and in vitro unveil a solvent-exposed hydrophobic pocket lined by acidic residues in CarS, where the CarA DNA recognition helix docks with high affinity in an atypical ligand-recognition mode for SH3 domains. Our findings uncover an unprecedented use of the SH3 domain-like fold for protein-protein recognition whereby an antirepressor mimics operator DNA in sequestering the repressor DNA recognition helix to activate transcription.
Organizational Affiliation:
Instituto de Química-Física Rocasolano, Consejo Superior de Investigaciones Científicas (CSIC), Madrid, Spain.