Hexammineruthenium(III) ion interactions with Z-DNA
Bharanidharan, D., Thiyagarajan, S., Gautham, N.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 1008-1013
- PubMed: 18084080
- DOI: https://doi.org/10.1107/S1744309107047781
- Primary Citation of Related Structures:
2HTO, 2HTT - PubMed Abstract:
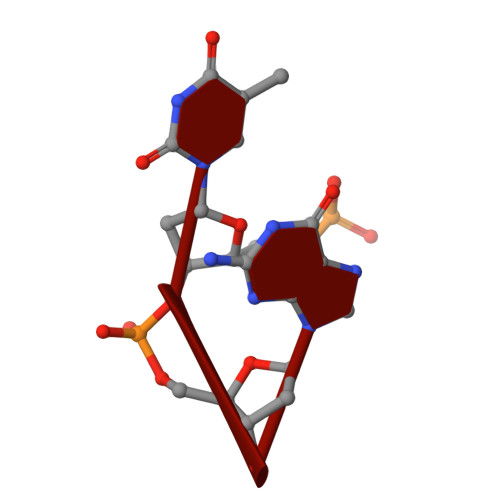
The hexamer duplex d(CGCGCA).d(TGCGCG) was crystallized with hexammineruthenium(III) ions in an orthorhombic space group; the crystals diffracted to 1.54 A resolution. Strong ion interactions with the adenine base induce a tautomeric shift from the amino to the imino form. Consequently, the A.T base pairing is disrupted. This structural study may be relevant to metal toxicity.
Organizational Affiliation:
Department of Crystallography and Biophysics, University of Madras, Guindy Campus, Chennai 600 025, Tamil Nadu, India.