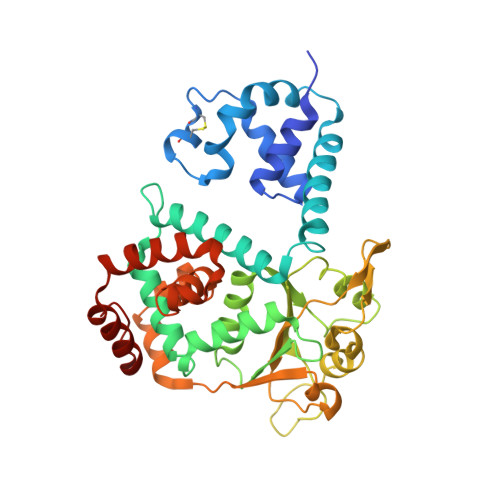
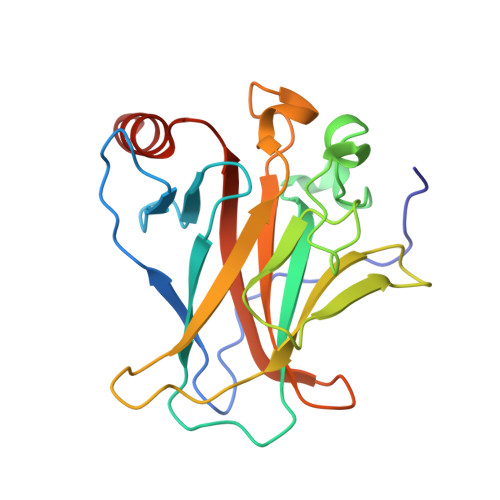
Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor.
Lilyestrom, W., Klein, M.G., Zhang, R., Joachimiak, A., Chen, X.S.(2006) Genes Dev 20: 2373-2382
- PubMed: 16951253
- DOI: https://doi.org/10.1101/gad.1456306
- Primary Citation of Related Structures:
2H1L - PubMed Abstract:
The transformation potential of Simian Virus 40 depends on the activities of large T-antigen (LTag), which interacts with several cellular tumor suppressors including the important "guardian" of the genome, p53. Inhibition of p53 function by LTag is necessary for both efficient viral replication and cellular transformation. We determined the crystal structure of LTag in complex with p53. The structure reveals an unexpected hexameric complex of LTag binding six p53 monomers. Structure-guided mutagenesis of LTag and p53 residues supported the p53-LTag interface defined by the complex structure. The structure also shows that LTag binding induces dramatic conformational changes at the DNA-binding area of p53, which is achieved partially through an unusual "methionine switch" within p53. In the complex structure, LTag occupies the whole p53 DNA-binding surface and likely interferes with formation of a functional p53 tetramer. In addition, we showed that p53 inhibited LTag helicase function through direct complex formation.
Organizational Affiliation:
Molecular and Computational Biology, University of Southern California at Los Angeles, Los Angeles, California 90089, USA.