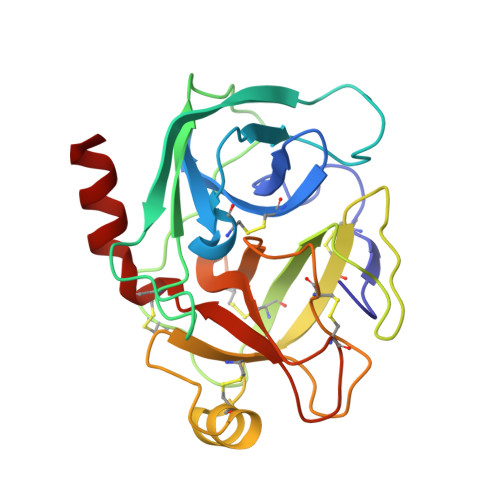
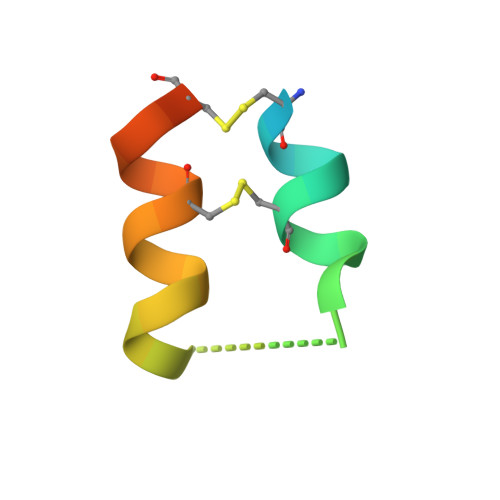
An Unusual Helix-Turn-Helix Protease Inhibitory Motif in a Novel Trypsin Inhibitor from Seeds of Veronica (Veronica Hederifolia L.).
Conners, R., Konarev, A., Forsyth, J., Lovegrove, A., Marsh, J., Joseph-Horne, T., Shewry, P., Brady, R.L.(2007) J Biol Chem 282: 27760
- PubMed: 17640870
- DOI: https://doi.org/10.1074/jbc.M703871200
- Primary Citation of Related Structures:
2CMY, 2PLX - PubMed Abstract:
The storage tissues of many plants contain protease inhibitors that are believed to play an important role in defending the plant from invasion by pests and pathogens. These proteinaceous inhibitor molecules belong to a number of structurally distinct families. We describe here the isolation, purification, initial inhibitory properties, and three-dimensional structure of a novel trypsin inhibitor from seeds of Veronica hederifolia (VhTI). The VhTI peptide inhibits trypsin with a submicromolar apparent K(i) and is expected to be specific for trypsin-like serine proteases. VhTI differs dramatically in structure from all previously described families of trypsin inhibitors, consisting of a helix-turn-helix motif, with the two alpha helices tightly associated by two disulfide bonds. Unusually, the crystallized complex is in the form of a stabilized acyl-enzyme intermediate with the scissile bond of the VhTI inhibitor cleaved and the resulting N-terminal portion of the inhibitor remaining attached to the trypsin catalytic serine 195 by an ester bond. A synthetic, truncated version of the VhTI peptide has also been produced and co-crystallized with trypsin but, surprisingly, is seen to be uncleaved and consequently forms a noncovalent complex with trypsin. The VhTI peptide shows that effective enzyme inhibitors can be constructed from simple helical motifs and provides a new scaffold on which to base the design of novel serine protease inhibitors.
Organizational Affiliation:
Department of Biochemistry, University of Bristol, Bristol BS8 1TD, United Kingdom.