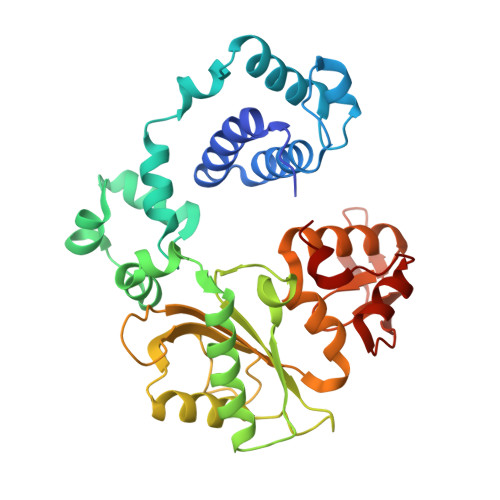
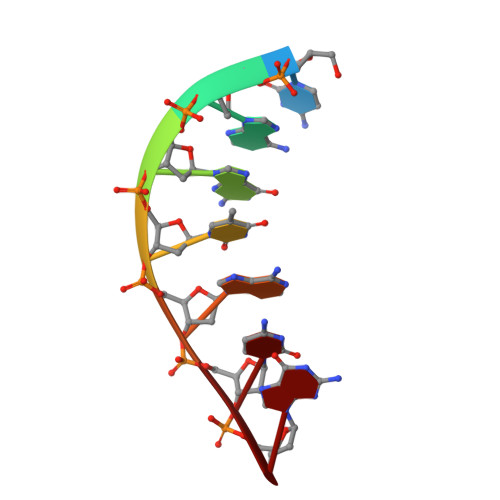
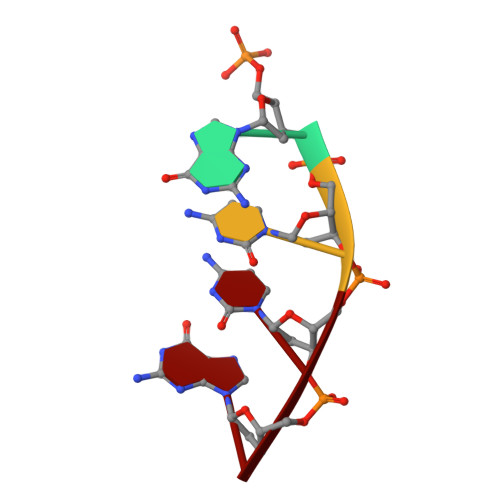
A closed conformation for the Pol lambda catalytic cycle.
Garcia-Diaz, M., Bebenek, K., Krahn, J.M., Kunkel, T.A., Pedersen, L.C.(2005) Nat Struct Mol Biol 12: 97-98
- PubMed: 15608652
- DOI: https://doi.org/10.1038/nsmb876
- Primary Citation of Related Structures:
1XSL, 1XSN, 1XSP - PubMed Abstract:
Pol lambda is a family X member believed to fill short gaps during DNA repair. Here we report crystal structures of Pol lambda representing three steps in filling a single-nucleotide gap. These structures indicate that, unlike other DNA polymerases, Pol lambda does not undergo large subdomain movements during catalysis, and they provide a clear characterization of the geometry and stereochemistry of the in-line nucleotidyl transfer reaction.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services, Research Triangle Park, North Carolina 27709, USA.